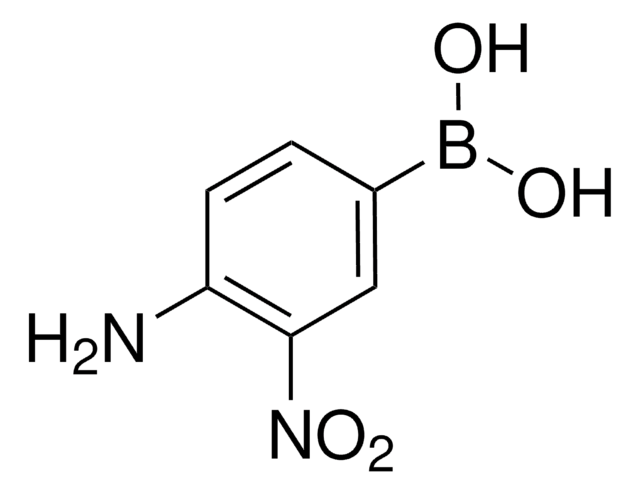
SRP0125
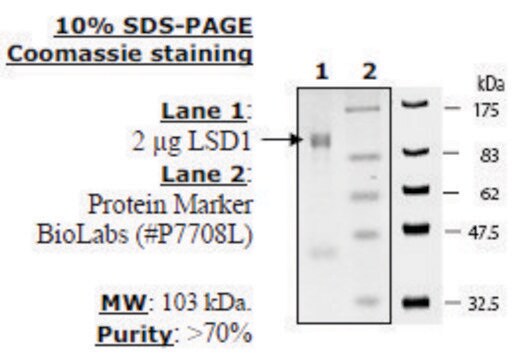
LSD1 substrate (Di-methylated K4_H3)
≥90% (HPLC), aqueous solution
Synonyme(s) :
HIST1H3E, Histone H3.1
About This Item
Produits recommandés
product name
LSD1 substrate (Di-methylated K4_H3), ≥90% (HPLC)
Source biologique
human
Pureté
≥90% (HPLC)
Forme
aqueous solution
Poids mol.
2283 Da
Conditionnement
pkg of 500UL
Concentration
0.55 mg/mL
Numéro d'accès UniProt
Conditions d'expédition
dry ice
Température de stockage
−70°C
Informations sur le gène
human ... HIST1H3E(8353)
Description générale
Application
Actions biochimiques/physiologiques
Forme physique
Notes préparatoires
Code de la classe de stockage
10 - Combustible liquids
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique