Products may be shipped at a different temperature than the recommended long-term storage temperature. If the product quality is sensitive to short-term exposure to conditions other than the recommended long-term storage, it will be shipped on wet or dry-ice. If the product quality is NOT affected by short-term exposure to conditions other than the recommended long-term storage, it will be shipped at ambient temperature. As shipping routes are configured for minimum transit times, shipping at ambient temperature helps control shipping costs for our customers. For more information, please refer to the Storage and Transport Conditions document: https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/marketing/global/documents/316/622/storage-transport-conditions-mk.pdf
C24604
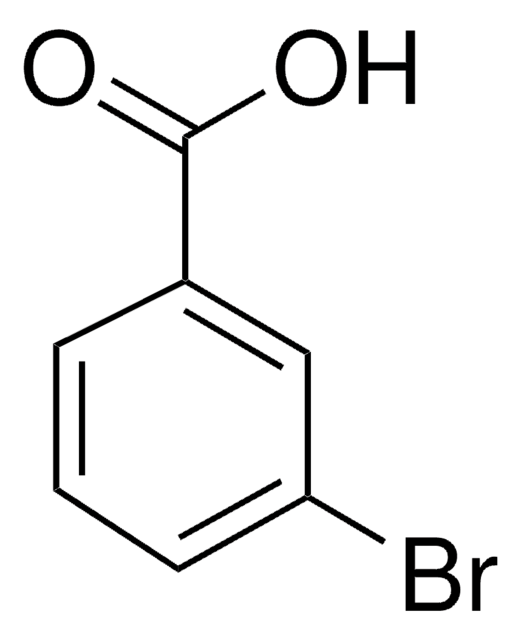
3-Chlorobenzoic acid
ReagentPlus®, ≥99%
Synonym(s):
m-Chlorobenzoic acid
About This Item
Recommended Products
Quality Level
product line
ReagentPlus®
Assay
≥99%
form
powder
mp
153-157 °C (lit.)
SMILES string
OC(=O)c1cccc(Cl)c1
InChI
1S/C7H5ClO2/c8-6-3-1-2-5(4-6)7(9)10/h1-4H,(H,9,10)
InChI key
LULAYUGMBFYYEX-UHFFFAOYSA-N
Looking for similar products? Visit Product Comparison Guide
Related Categories
Legal Information
Signal Word
Warning
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2 - Skin Irrit. 2
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Personal Protective Equipment
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
-
How is shipping temperature determined? And how is it related to the product storage temperature?
1 answer-
Helpful?
-
-
How can I determine the shelf life / expiration / retest date of this product?
1 answer-
If this product has an expiration or retest date, it will be shown on the Certificate of Analysis (COA, CofA). If there is no retest or expiration date listed on the product's COA, we do not have suitable stability data to determine a shelf life. For these products, the only date on the COA will be the release date; a retest, expiration, or use-by-date will not be displayed.
For all products, we recommend handling per defined conditions as printed in our product literature and website product descriptions. We recommend that products should be routinely inspected by customers to ensure they perform as expected.
For products without retest or expiration dates, our standard warranty of 1 year from the date of shipment is applicable.
For more information, please refer to the Product Dating Information document: https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/marketing/global/documents/449/386/product-dating-information-mk.pdfHelpful?
-
Active Filters
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service