Protein Fingerprinting of a Viral Vector, AAV5
Geoffrey Rule, Kevin Ray, Uma Sreenivasan, Cory Muraco, Pei Liu
Merck

Introduction
The culture and use of adeno-associated virus (AAV) as a gene delivery device has seen much interest in recent years as a strategy for delivering targeted gene therapies towards muscle, nerve, liver, and eye disorders. This includes different AAV serotypes that target specific tissues as well as genetically engineered hybrid types with altered tissue specificity.
In-depth characterization of the viral capsid proteins, as well as the genomic content, is essential to verify critical quality attributes of these particles. Both the amino acid sequence as well as post-translational modifications (PTMs) that are found on the viral capsid proteins can have an impact on the tissue tropism, efficacy, and immunogenicity of AAV.1 PTMs that have been identified on AAV include phosphorylation, SUMOylation, ubiquitination, acetylation, methylation, and glycosylation.2
The USP released draft guidelines3 for the analytical characterization of viral vectors in 2022 to provide method starting points for determination of critical quality attributes. A variety of tests are described to characterize the identity, purity, concentration, and potency of these viral vectors, among other traits. For determination of capsid identity, starting methods are provided that use Western blotting, reversed-phase HPLC with UV detection, and HPLC with MS detection. The latter approach describes the use of both intact mass analysis of the capsid proteins, as well as amino acid sequence analysis using peptide mapping. Here we describe our work to develop methods for protein fingerprinting of AAV serotype 5 using both intact mass analysis and peptide mapping. Several post- translational modifications of the viral proteins VP1, VP2 and VP3 were identified.
Experimental Methods
A system suitability mix, MSRT1, was prepared according to the instructions on the data sheet but with a final acetonitrile concentration of 1.6% (v/v). The injection volume was 10 µL. This solution is a mix of 14 isotopically labelled peptides injected prior to injection of samples to verify instrument performance.
AAV Production
AAV5 subtype (Q9YIJ1) was produced in HEK293 non-T VirusExpress® 293 AAV Production Cells using EX-CELL® CD HEK293 Viral Vector Medium. Three transfection plasmids carrying: 1. Replication and capsid genes; 2. certain adenovirus genes on a helper plasmid and 3. a plasmid with the gene of interest, in this case green fluorescence protein (GFP), were used. After three days of transfection, full AAV capsids, as well as product related impurities such as empty capsids and capsids filled with host DNA fragments or plasmid backbones, were produced. The HEK cells were then lysed with detergent, treated with benzonase/magnesium, to digest all unused plasmid DNA and host nucleic acids, and finally treated with 0.5 M sodium chloride. The clarified lysate was affinity purified on a fast protein liquid chromatography (FPLC) system (ÄKTA) using commercially available resin, from host cell proteins and digested nucleic acids. This process resulted in viral particle (vp) concentrations between 4E+12 and 2E+13 vp/mL as determined by droplet digital polymerase chain reaction (ddPCR).
Intact Mass Analysis
AAV particles were treated with 10% acetic acid for 15 minutes to dissociate AAV capsid assembly before centrifugation at 12,000 rpm for five minutes. Approximately 0.6 µg of total AAV capsid protein was injected for each run.
Chromatography was performed on BIOshell™ A400 Protein C4, 3.4 µm particle, and BIOshell™ IgG 1000 Å C4, 2.7 µm particle columns, along with a competitor column, mentioned in the draft guideline, for comparison. All had dimensions of 100 x 2.1 mm. Mass spectrometry was conducted on a Waters™ Xevo™ G2-S QTof in positive ion mode. The chromatographic conditions and instrument parameters used for the applied LC-UV-MS method are shown in Tables 1 and 2 below.
Peptide Mapping
Digestion of purified AAV particles was conducted with both trypsin and, separately, chymotrypsin, using a Low-Artifact Digestion Buffer and a filter-assisted, sample preparation protocol. For details on this protocol, please see the online application note.4
BIOshell™ A160 Peptide C18, 2.7 µm particle column and an identical BIOshell™ column with 2 µm particles were used with gradient conditions outlined in the USP draft guidelines.3 Both columns were 150 x 2.1 mm in dimension. For comparison, a competitor C18 column, mentioned in the draft guidelines, was evaluated using the same conditions (Table 3) and column dimensions.
Mass spectrometry was conducted on a Thermo QE Plus in positive ion mode using a scan range of 200 to 2000 m/z and data dependent MS2 of the top ten ions (Table 4).
Results
Protein Fingerprinting
The draft USP guidelines provided a starting point for MS characterization of viral vectors by both intact capsid fingerprinting and peptide mapping. The draft guideline also describes an additional method, using UV detection and a 2-hour chromatographic run, for determination of capsid stoichiometry. While we did not replicate this method, we did use UV detection in conjunction with mass spectrometry fingerprinting of the intact viral capsids over a shorter 30-minute run. Integration of the UV detected peaks was then used to evaluate capsid stoichiometry. The combined LC- UV-MS analysis provided a convenient, one-method assessment of fingerprint and stoichiometry in a shorter run time.
Intact Mass Analysis of VP1, VP2, and VP3
The comparison of columns for separation of the intact capsid proteins is shown in Figure 1. Both the BIOshell™ A400 C4 and BIOshell™ IgG 1000 Å C4 columns provided good retention of the proteins using the gradient described in the draft guideline. Separation of VP1 from VP2 and VP3 was also achieved. The BIOshell™ A400 C4 column also provided partial separation of a VP3-clip from VP3. The competitor column gave faster elution with the same conditions but did not show a distinct peak for VP1. Deconvolution of the peaks observed on the BIOshell™ A400 C4 column resulted in the mass determinations as shown in Figure 2.
Both VP1 and VP3 were observed to be highly acetylated while VP2 showed a degree of phosphorylation. The presence of 0.1% TFA in the mobile phases was found to result in a substantial amount of TFA adduction on each of the proteins. Interestingly, other AAV serotypes (AAV2, AAV8) we have examined have not shown this same degree of TFA adduction. Replacing the TFA with 0.1% formic acid resulted in a loss of chromatographic separation of the three capsid proteins although the TFA adduction was eliminated (data not shown). The use of difluoroacetic acid (DFA) could be evaluated as a compromise between chromatographic separation and MS sensitivity.
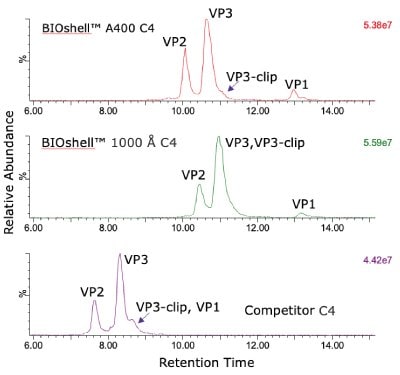
Figure 1.Total ion current (TIC) profile comparison of AAV5 capsid protein retention and separation on three columns evaluated.
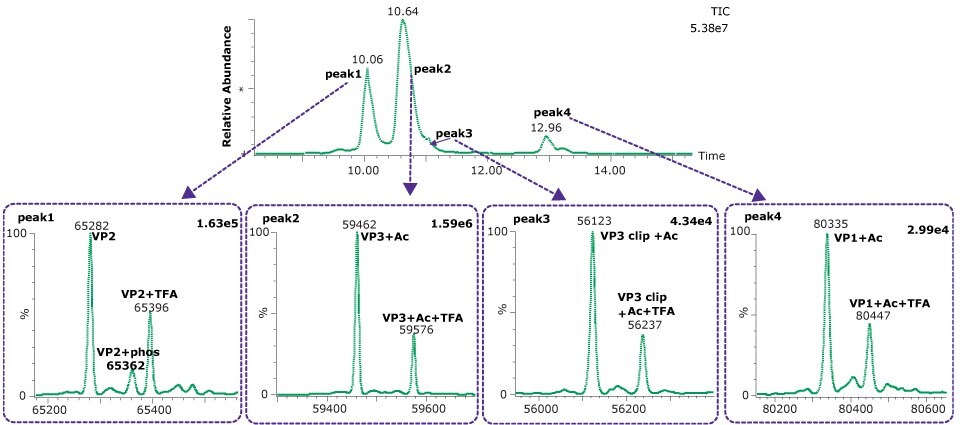
Figure 2.Deconvoluted spectra from each of four dominant peaks observed on the BIOshell™ A400 Protein C4 column.
A clipped form of VP3 was also observed, corresponding to the cleavage of the aspartic acid (695) - proline (696). This bond is known to be particularly labile to hydrolysis under acidic conditions and elevated temperature, and similar clip proteoforms have been reported by others.5,6 Table 5 shows the close agreement of the observed masses and the theoretical masses for the capsid proteins.
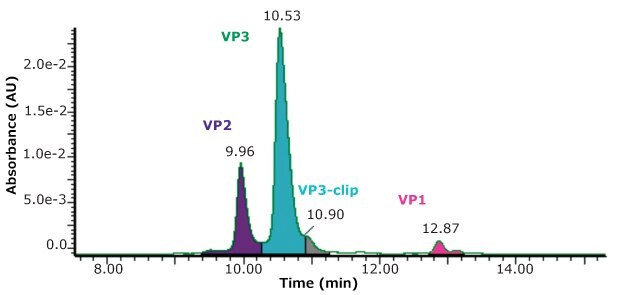
Using the same conditions described above, but with UV detection, a measurement of the relative abundance of the four main peaks was made, as shown in Figure 3.
While the stoichiometry of AAV capsids is sometimes suggested to have a specific ratio of capsid proteins, such as 1:1:10, Wörner et al.7 nicely describe the process of capsid formation as being a stochastic sampling of capsid proteins available in the cell pool. AAV capsids are then made up of a highly heterogenous composition of three capsid proteins so that measured stoichiometry represents only an average composition. Combining both the UV and MS detection in the same run is an added convenience in the evaluation of samples.
Peptide Mapping
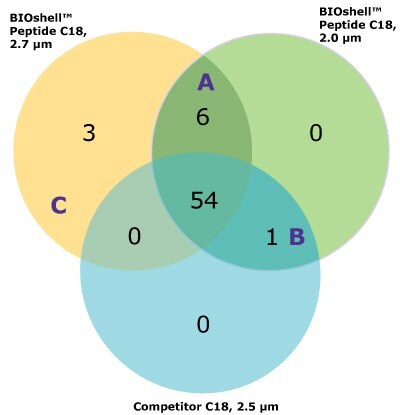
For the comparison here, we used the column and conditions suggested in the draft USP guidelines; a competitor column and gradient conditions shown in Tables 3 and 4, above. This gradient is delivered over a 50-minute period. On both the competitor and BIOshell™ columns, good sequence coverage was obtained using tryptic digestion but with slightly better sequence coverage on the BIOshell™ columns due to retention of several small, early eluting peptides including VVTK, ADEVAR, GEPVNR, SLRVK, RIDDHFPKR (Figure 4). Importantly, the N-terminal sequence of VP2, (APTGK) was also identified on these columns, in addition to the N-terminal sequences of VP1 and VP3 as shown in Figure 5.
A |
|---|
B |
|---|
C |
|---|




Figure 5.Product ion spectra providing sequence coverage of the N-termini of VP1 (A), VP2 (B), and VP3 (C) as well as C-terminus of each (D).
One large section of 60 amino acids was not covered by tryptic digestion alone, and so a separate digestion was performed with chymotrypsin to increase the overall sequence coverage. Similar multi-enzyme digestion of AAV capsids has been shown useful by others.8 Using the two enzymes, separately, increased the coverage to 100% on the BIOshell™ columns.
A summary of the PTMs identified, along with their percent abundance on each of the three columns, is shown in Table 6.
Both VP1 and VP3 were found to be 100% acetylated while at least four sites of phosphorylation were identified. Acetylation of the N-terminus of AAV capsids appears to be highly conserved across serotypes.2 In addition to the deamination site, four sites of oxidation were observed with all being less than 1% abundant than the unoxidized forms. Overall, the agreement in the abundances of the PTMs between the columns was very good.
Conclusions
Several column comparisons were shown to demonstrate uses of the BIOshell™ line of columns for characterizing viral vectors, in this case AAV serotype 5. Conditions outlined in the USP draft guideline were used for both intact mass fingerprinting of viral capsids and for peptide mapping experiments, but we suggest that further improvements in chromatography might be made with additional gradient optimization.
The BIOshell™ columns, particularly the A400 Protein C4 column, have proven to be effective in separating the three capsid proteins of AAV5 for intact mass analysis and stoichiometry evaluation. In addition, the BIOshell™ A400 Protein C4 column provides partial separation of the VP3- clip from VP3. The competitor column also shows partial separation of VP3-clip from VP3 but with coelution of VP1.
The BIOshell™ A160 Peptide C18 columns, in both the 2.7 and 2.0 µm particle sizes, proved to be useful in retaining short, polar peptides to provide slightly improved sequence coverage over the competitor column. Retention of the N-terminus of VP2 was only provided by the BIOshell™ columns.
The USP draft conditions for the mobile phase and gradient conditions were used in both approaches for characterizing capsids, but we suggest these conditions might benefit from further optimization with other AAV serotypes or specific PTMs.
Acknowledgements
The authors want to thank Dr. Lass-Napiorkowska of Merck, Process Solutions R&D, Upstream BioProcessing, St. Louis, MO, for providing the AAV5 serotype
References
To continue reading please sign in or create an account.
Don't Have An Account?