An LC/MS Peptide Standard for Rapid System Suitability Assessment and Retention Time Translation Across LC/MS Platforms
Kevin Ray, Jim Walters
Reporter US Volume 34.2
Introduction
Standards are critical in liquid chromatography/mass spectrometry (LC/MS) based proteomics to ensure optimal and consistent system performance before, during, and after sample analysis. They may be used to assess peptide elution, to troubleshoot chromatography, to demonstrate that the LC/MS platforms are working properly, and to translate retention times across platforms. Routine platform suitability checks and standardization are required in order to relate experimental data across and between analytical runs. It is especially critical when transferring assays or comparing data across platforms or laboratories. These requirements demand a well-developed and characterized standard. The objective of this project was to develop a mixture of synthetic peptides designed to act as an LC/MS platform standard, containing elements to test LC resolution, peptide elution profile, and overall LC/MS platform performance as well as to enable retention time translation across platforms.
Read more about
Experimental
We screened a library of peptides to identify a set with good chromatographic peak shape and MS intensity across a broad range of hydrophobicities designed to span the normal elution profile of complex proteomic samples. Other design considerations included avoidance of any potential interference with target peptides of interest, and the ability to allow for a quick visual inspection of easily resolved components with similar peak intensities (based on electrospray response). The result is a mixture of 14 stable isotope labeled (SIL) peptides (Table 1) for performing a quick, robust test of platform suitability, cross platform comparison, and retention time translation. The peptide mix (MSRT1) is routinely injected on all LC/MS proteomic platforms in our laboratory for routine tracking of instrument performance.
Amino acid in brackets denotes site of label incorporation as follows: [K],13C6 15N2; [R],13C6 15N4; or [L],13C6 15N1
Further, we have established universal retention factors, or so-called iRT’s,1 for each peptide in the MSRT1 standard. Subsequently, retention factors were established for target peptides of interest from a discovery proteomics sample set relative to the reference MSRT1 peptides. With a single calibration run of MSRT1 on another LC/MS platform intended for targeted quantitation of peptides, these retention factors were used to translate the retention time of the target peptides under the new conditions and schedule MRM acquisition windows.
Results and Discussion
As shown in Figure 1, using MSRT1, all 14 peptides are readily resolved and observed, indicating an operational analytical platform for sample analysis. Multiple parameters from this daily check can be monitored over time in the form of control charts tracking injections performed over several months. We have successfully used these metrics to quickly identify instrument performance problems such as retention time shifts and longitudinal drifts in mass accuracy as shown in Figure 2.
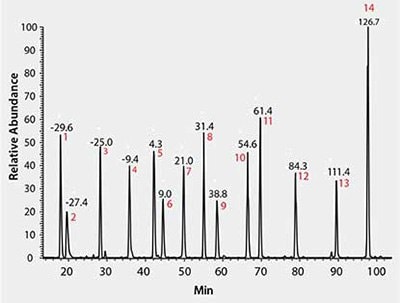
Figure 1.LC/MS Total Ion Chromatogram of MSRT1
![control-chart-mass-accuracy Statistical Control Charts for Mass Accuracy and Retention Time of Peptide LVNEVTEFA[K]](/deepweb/assets/sigmaaldrich/marketing/global/images/technical-documents/articles/analytical-chemistry/large-molecule-hplc/control-chart-mass-accuracy/control-chart-mass-accuracy.jpg)
Figure 2.Statistical Control Charts for Mass Accuracy and Retention Time of Peptide LVNEVTEFA[K]
We also utilize the mixture to compare proteomic platform performance and characteristics, such has diagnosing potential system biases against either early or late eluting peptides. For example, peptides 1 and 2 of MSRT1 are not readily observed on various sample trapping columns and conditions indicating very hydrophilic peptides in our samples will also being lost and not detected under these conditions. Using MSRT1 we have been able to optimize trapping chemistry, volume, and flow rate to achieve detection of early eluting peptides for applications in which this is required. This highlights the utility of a standard which mimics the span of the typical elution profile of a complex proteomic sample and allows platform optimization for samples, without actually consuming precious samples.
Finally, we implemented a method using MSRT1 to translate retention times of peptide targets across instrument platforms. We have established iRT’s for all the peptides in MSRT1 (peak labels in Table 1) using a proteomics discovery workflow. Subsequently, only one calibration run on a quantitative platform is required to convert the iRT’s into a translated retention time on this, or any, new method or LC/MS platform (Figures 3 and 4). This is accomplished by spiking MSRT1 peptides into the sample and the retention times of the target peptides are predicted using the linear equation generated by the retention time and iRT’s of the MSRT1 peptides, aided by the free software, Skyline.2 This makes it possible to translate the stable retention property of a peptide into accurate prediction of retention time on other LC set-ups (Figure 5) leading to higher multiplexing and improved peak picking and scoring in MRM analysis. Additionally, in MRM assay transfer and comparison, time and resources can be maximized when peptide target retention times can be accurately translated across platforms in this manner.

Figure 3.Measured Retention Times of MSRT1 Peptides Versus Retention Factor

Figure 4.Illustration of the Retention Time Translation of Five Peptides of Interest

Figure 5.Plot of the Observed Retention Times
Conclusions
A 14 SIL peptide mix, MSRT1, has been developed for daily assessment of LC/MS based proteomic platforms. This mixture allows for a quick check of system suitability and tracking critical parameters over time. In addition, MSRT1 mimics and spans the typical elution profile of a complex proteomic mix allowing for platform optimization prior to sample injection. Finally, MSRT1 can be used to translate retention times across instrument platforms and between laboratories.
References
To continue reading please sign in or create an account.
Don't Have An Account?