Mass Spectrometry of Glycans
The development of modern mass spectrometry (MS) equipment with high resolution and mass accuracy has led to its use in analyzing glycans for both profiling and structural studies in glycomics and glycoproteomics research. Unlike the HPLC method, a substantially larger amount (~10‑20 times) of glycan is normally required for a single MS spectrum. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry is the most widely used MS technique. The information gained by using MALDI-MS is mass weight, which can be used to assign putative monosaccharide structures present in a pure oligosaccharide since the mass of a monosaccharide is measured with a high degree of accuracy. MALDI-MS can be used in conjunction with glycan sequencing using exoglycosidase enzymes for structural and linkage analysis of glycans.
Different approaches and matrices are used for the analysis of neutral and acidic glycans (those that contain sialic acid) by MALDI-MS. Neutral glycans ionize with reasonable efficiency using 2,5‑dihydroxybenzoic acid (2,5‑DHB) as the matrix in positive ion mode. Acidic glycans generally provide poor MALDI spectra with DHB matrices due to variable losses of sialic acids or carboxyl groups, leading to multiple peaks. These glycans are therefore analyzed in negative ion mode using an alternative matrix as described later.
Read more about
Mass Spectrometry of Neutral Glycans
Unlike peptides, neutral glycans exhibit low ionization efficiency and the [M+H]+ ion is therefore not sufficiently abundant. However, these glycans can be detected as alkali metal adducts which ionize efficiently. In most procedures, [M+Na]+ is the major ion, accompanied by a weaker [M+K]+ ion. Other adducts, such as [M+Li]+ can be generated by the addition of the appropriate inorganic salt to the matrix. The inclusion of NaCl in the matrix solvent allows the glycans to ionize predominantly in the [M+Na]+ form with little or no [M+K]+ ion formation.
Normally, use of the 2,5‑DHB matrix is sufficient for most applications. However, it has been reported that, in some instances, a 2‑3 fold increase in sensitivity can be obtained with the use of a "Super DHB" matrix as it allows for “softer” desorption with a reduction in metastable ion formation. "Super DHB" consists of a mixture of 2,5‑DHB and 2‑hydroxy-5‑methoxybenzoic acid (90:10 ratio, wt %).
Ovalbumin (0.5 mg) was incubated with 7 units of proteomics grade PNGase F enzyme at 37 °C for 3 hours and the glycans were recovered by ultrafiltration. A third of this material was dried under vacuum centrifugation and then analyzed by MALDI-TOF MS. Figure 1 shows the N-linked glycan profile of ovalbumin, with identification assignments made to twelve of the signals (Figure 2).
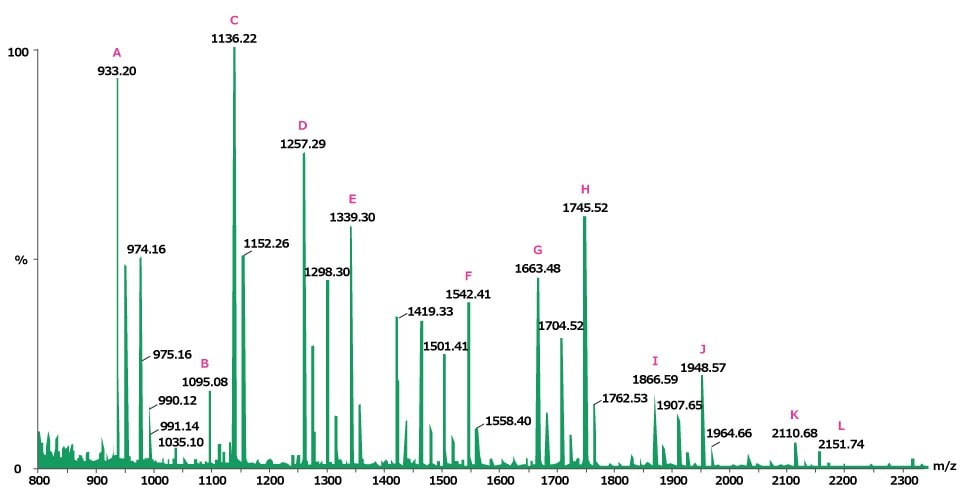
Figure 1.Positive ion MALDI-MS of the neutral N-linked glycans from ovalbumin released by treatment with PNGase F and recorded using 2,5‑DHB as the matrix. The letters (A-L) indicate identified peak structures, as shown in Figure 2.
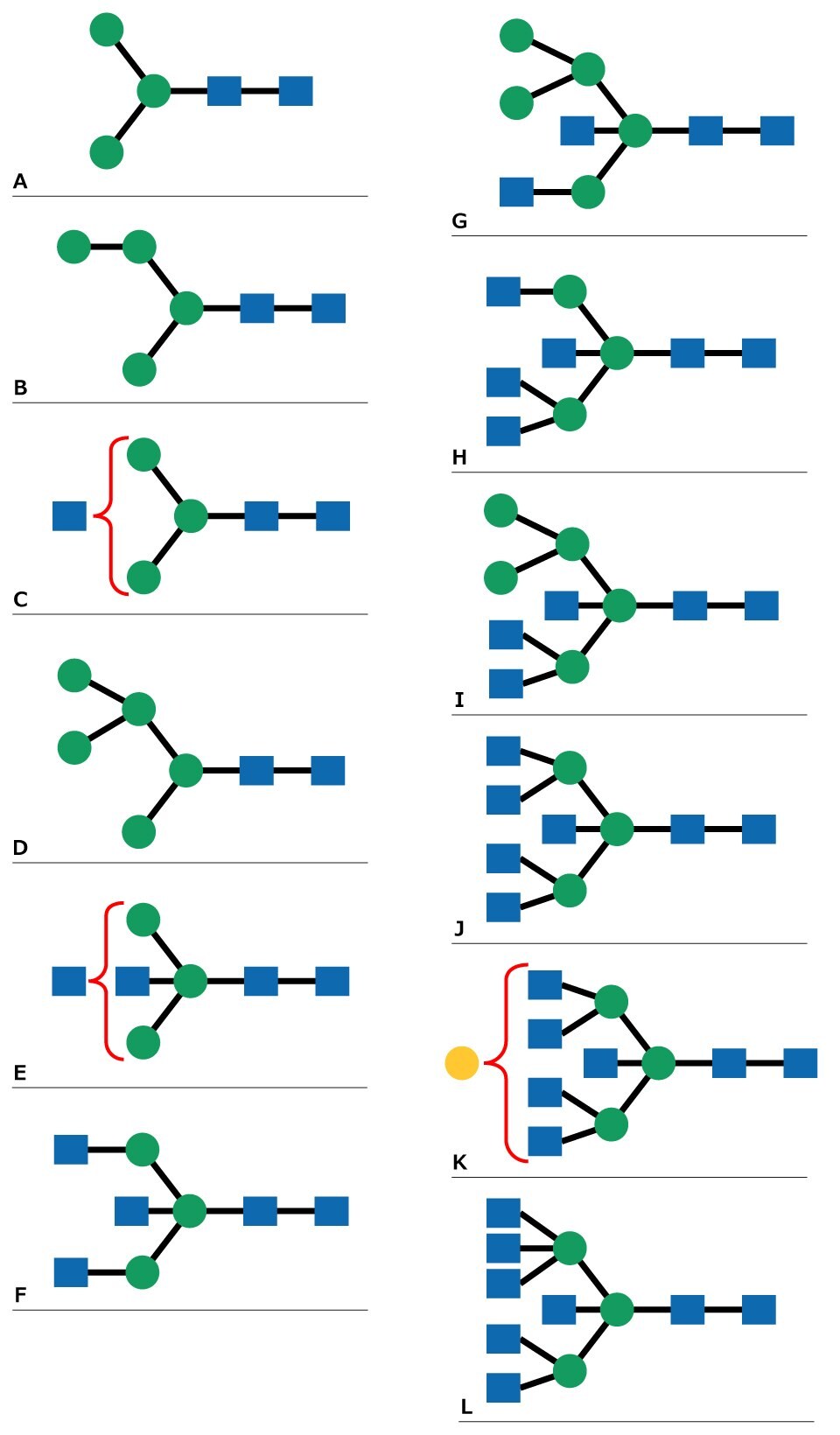
Figure 2.N-Linked glycan structures that correspond to MALDI MS peaks in Figure 1.
General Mass Spectrometry Procedure for Glycan Analysis
The following method, commonly referred to as the dried-droplet method, is based on the original MALDI experiments and remains the most commonly used method in the mass spectrometry community.
- Transfer 3 μL of the appropriate matrix solution (either DHB or "Super DHB" at 10 mg/mL) into a 0.5 mL Eppendorf® tube.
- Add 1 μL of 10 pmol/μL solution of a standard glycan or an unknown sample solution to the tube containing the matrix; vortex to mix.
- Dispense an aliquot (about 1.5 μL) of this mixture onto the MALDI target plate.
- Allow to dry at room temperature.
Note: Mass spectra can be acquired at this stage. However the following steps have been reported to improve signals by forming an even film of crystals, which reduces the need to search for “sweet spots” on the target.
- Onto the dried spot, dispense 1 μL of ethanol and allow the matrix plus analyte mixture to recrystallize on the target plate.
- Dry the spot. The target is now ready to acquire mass spectra.
Figure 3 shows representative MALDI-TOF MS traces obtained for three glycan standards Man-3 glycan (A), Man-8 glycan (B), and NA4 glycan (C).
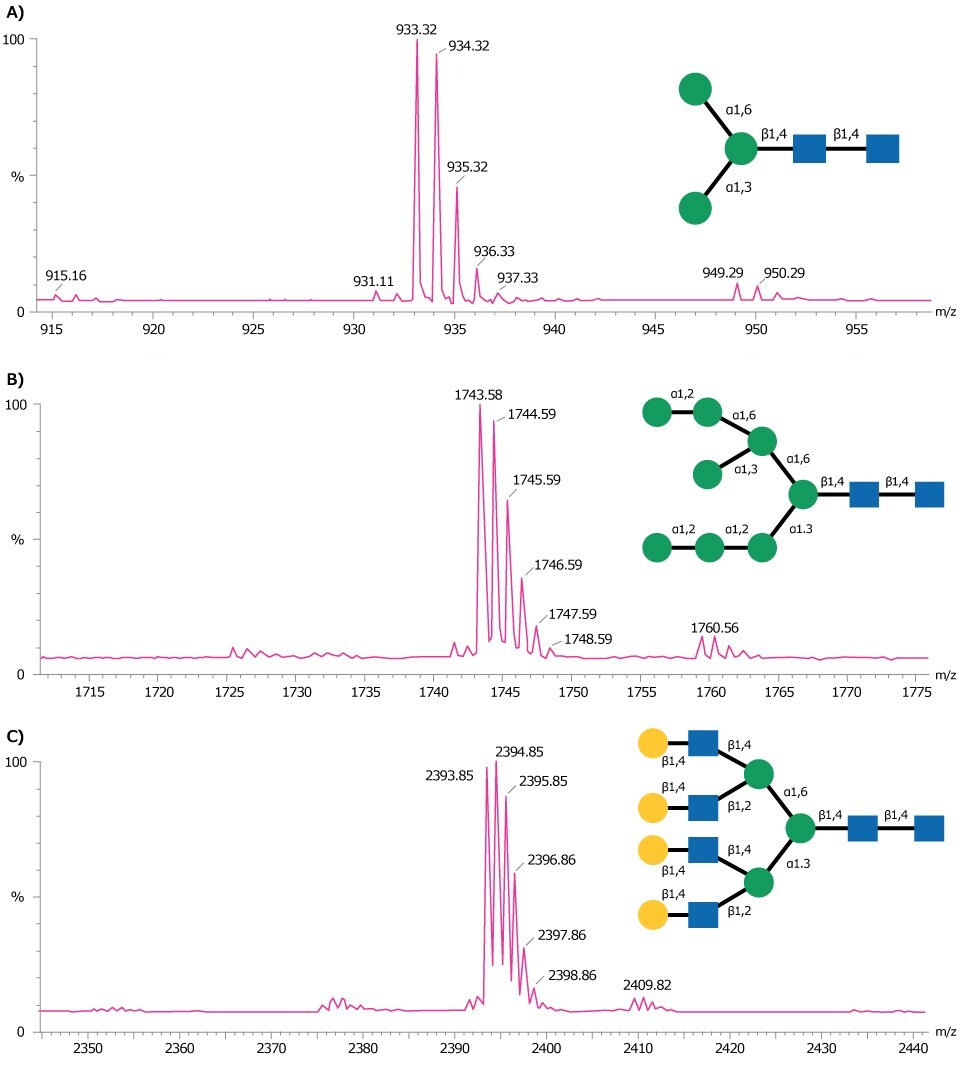
Figure 3.MALDI-TOF mass spectra of (A) Man-3 glycan, (B) Man-8 glycan, and (C) NA4 glycan obtained in positive ion reflectron mode using DHB as matrix. Each of these glycans are detected as the sodium adduct of the molecular ion [M+Na]+.
Download an app note on a workflow for released N-glycan analysis.
Mass Spectrometry of Acidic Glycans
DHB is the most widely used matrix for the MALDI-MS analysis of glycans. This matrix, however, is not ideal for acidic glycans (such as sialylated N-linked sugars) as the detection limit is poor compared to neutral glycans, and fragmentation can occur with losses of carboxylic and/or sialic acid groups. In addition, sialylated glycans analyzed in the positive-ion mode yield a mixture of cation adducts producing multiple peaks. One approach to enhance sensitivity is to derivatize the glycan, but this may compromise the absolute quantitation of the glycan. An alternative way is to choose a different matrix that can overcome the problem of fragmentation during analysis and an increased detection limit.
Two widely used matrices for the analysis of acidic glycans are 6‑aza-2‑thiothymine and 2,4,6‑trihydroxyacetophenone (THAP). The former gives a significant increase in sensitivity over DHB matrix, but still causes some fragmentation in the negative ion linear mode. With THAP, the detection limit of analysis can be at the 10 fmol level, with little or no evidence of fragmentation in the linear mode. The conditions outlined below for sample preparation with this matrix are crucial for obtaining maximum sensitivity.
In order to prevent acid-catalyzed lactonization of sialic acids, glycans were dissolved in 0.2 M ammonium hydroxide solution before being applied to the MALDI target plate and air dried. The spot was re-hydrated with water and a mixture of THAP and ammonium citrate (1 mg THAP in a 1 mL mixture of 1:1 acetonitrile:20 mM ammonium citrate) was applied. Vacuum drying of the sample spot was used to prevent large crystals from forming. The sample was then allowed to absorb moisture from the atmosphere to promote the formation of small crystals.
Figure 4 is representative of a MALDI-TOF MS spectrum obtained in negative ion linear mode for the monosialylated N-linked glycan A1.
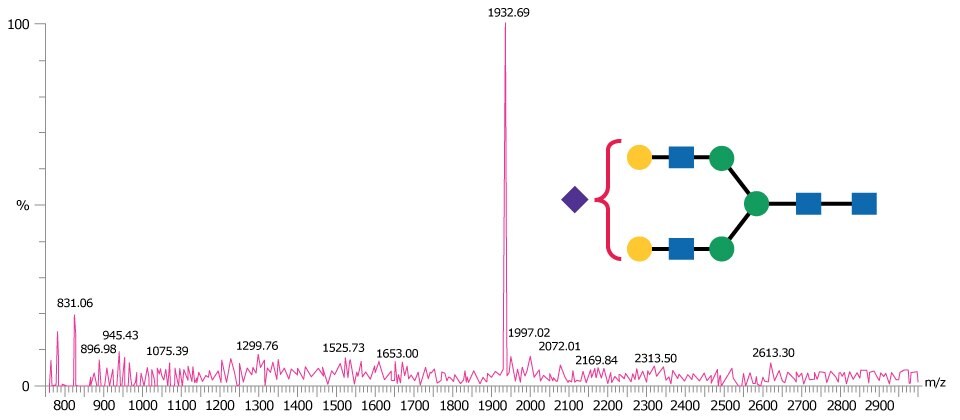
Figure 4.MALDI-TOF mass spectrum of A1 glycan obtained in negative ion linear mode using THAP as matrix and detected as the [M-H]- ion.
Find more technical resources on our Glycobiology hub page or our Protein Mass Spec hub page.
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?