Glycan Labeling
Glycobiology Analysis Manual, 2nd Edition
GlycoProfile™ Products for Metabolic Glycan Labeling
Structural modifications of proteins are essential to living cells. When aberrantly regulated they are often the basis of disease. Glycans are responsible for much of the structural variation in biologic systems, and their representation on cell surfaces is commonly called the “glycome.” Researchers are beginning to see the potential for breakthrough in healthcare through glycomics, which studies carbohydrates, proteins, glycoproteins, and their interactions.
- Bio-Orthogonal tracking of glycan biosynthesis
- Use the power of FLAG® for detection and enrichment
We offer tools for rapid profiling of N and O-linked glycoproteins by metabolic labeling with N-azidoacetylgalactosamine, acetylated (Gal-NAz), N-azidoacetylglucosamine, acetylated (GlcNAz), and/or N-azidoacetylmannosamine, acetylated (ManNAz) followed by modified-Staudinger ligation with FLAG-phosphine probes. Glycoproteins are robustly labeled or enriched with ANTI-FLAG® conjugates permitting analysis of the glycosylation machinery or metabolism. Metabolic labeling followed by modified-Staudinger ligation provides a means for proteomic analysis of this post-translational modification and identifying glycoprotein fingerprints associated with disease.
Cells metabolize the azidosugars using glycosyltransferases and express the sugars on the glycan chain both intracellularly and on the cell surface, leaving the azido group unreacted. The azidosugars can also be incorporated into glycans via the sialic acid metabolic pathway. A selective phosphine probe containing the detection FLAG epitope is applied to the cell or extracts containing the azidoglycans. The phosphine group selectively reacts via Staudinger ligation with the displayed azido group, resulting in an epitope tag covalently attached to the glycan.
Although non-natural molecules, both the azido and phosphine moieties are tolerated during cell proliferation. The bound epitope peptide is then detected by using a FLAG-specific antibody. This approach permits the analysis of pathways that are regulated by particular glycan post-translational modifications as well as the monitoring of the intracellular glycosylation process itself. Metabolic labeling with bio-orthogonal chemical reporters such as azidosugars followed by Staudinger ligation provides a unique mechanism for proteomic analysis of this post-translational modification and for identifying glycoprotein fingerprints associated with disease.
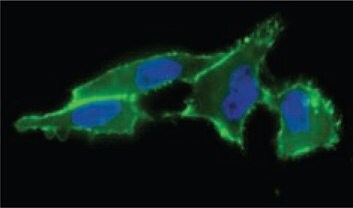
Figure 1. Cell surface sialic acid presentation detected with ANTI-FLAG®-FITC (Cat. No. F4049) (in green) showing where FLAG-Phosphine reacted with incorporated N-azidoacetyl-mannosamine, acetylated.

Figure 2. Profiling O-type glycoproteins by metabolic labeling with an azido GalNAc analog (GalNAz) followed by Staudinger ligation with a phosphine probe (FLAGphosphine).
References
GlycoProfile™ Labeling Kits
Useful Fluorescent Dyes for Enhanced Glycan Analysis
2‑AA and 2‑AB Labeling of Glycans by Reductive Amination
The GlycoProfile 2‑AA and GlycoProfile 2‑AB Labeling Kits contain reagents for labeling glycans at their reducing ends by reductive amination. The fluorophores 2‑AA (anthranilic acid) and 2‑AB (2‑aminobenzamide) provide valuable tools for glycan analysis due to their sensitivity and stability when coupled to glycans. Other commonly used methods, such as radioisotopic labels, antibody labels, and various probes do not display the stability, flexibility, and ease of use observed with 2‑AA and 2‑AB.
The glycans are labeled in a two-stage reductive amination process. Glycans with a free reducing sugar are in equilibrium between the cyclic or closed ring and the acyclic or open ring structure. In the first step, a stable Schiff's base is formed when the carbonyl carbon of the acyclic reducing sugar is attacked by the dye in a nucleophilic manner. Following the formation of the Schiff's base, the resulting imine group is reduced using sodium cyanoborohydride to yield a stable labeled glycan.
Labeling using 2‑AA / 2‑AB can be performed on either purified or pooled samples, including a variety of sources, such as N-linked, O-linked, and GPI anchored glycans. For samples containing sialylated oligosaccharides, sialic acid loss is negligible. Samples containing mixed pools of glycans can often be detected at femtomolar concentrations. Glycan binding to both dyes is very robust with no degradation during post-cleanup analysis.

Figure 3. Acyclic glycan and dye form a Schiff's base. Subsequent reduction of the imine with sodium cyanoborohydride results in a stable labeled glycan. (A) 2‑AA fluorophore (B) 2‑AB fluorophore.
Analysis of 2‑AA and 2‑AB Labeled Glycans
Once the glycans have been labeled, a variety of methods exist to analyze them. The most common techniques employ fluorescent detection after separation by HPLC or CE. These include separation by ion exchange, normal phase/RP HPLC, or size exclusion chromatography.
The labeled glycans are undetected by UV detection, but significant peaks are seen by fluorescence. The different chromatograms are due to the labeling efficiency, sensitivity, and other dye properties. Neither UV nor fluorescent detection was able to detect unlabeled fetuin glycans (data not shown). Labeled glycans can also be detected using mass spectrometry. Mass spectrometry can be performed with either an electrospray ionization (ESI) or matrix assisted laser desorption ionization (MALDI) ion source. Samples containing mixed pools of glycans can often be detected at picomolar concentrations.

Figure 4. HPLC profile of the 2‑AA labeled N-linked glycan library obtained from bovine fetuin. The glycans were cleaved from the glycoprotein using the Enzymatic Protein Deglycosylation Kit (Cat. No. EDEGLY).

Figure 5. HPLC profile of the 2‑AB labeled N-linked glycan library obtained from bovine fetuin. The glycans were cleaved from the glycoprotein using the Enzymatic Protein Deglycosylation Kit (Cat. No. EDEGLY).
GlycoProfile™ 2‑AA and 2‑AB Labeling Kits
Each GlycoProfile™ Labeling Kit contains sufficient reagents for labeling up to 36 samples. Two sets of components have been provided; each set is sufficient for labeling up to 18 samples based on a reaction volume of 5 μL. Mixed glycan samples should contain between 100 picomoles to 50 nanomoles of purified glycans. With a single pure glycan, as little as 5 picomoles may be labeled and detected in subsequent HPLC analysis.
Additional Labeling Reagents
Chemical derivatization is the most common method used for labeling glycans at their reducing ends by reductive amination. A single molecule of fluorescent label can be incorporated to each mono- or oligosaccharide. The sensitivity of detection by this technique is in the low femtomole range and depending upon the analytical technique, an appropriate fluorescent label can be used. Table 1 lists some of the fluorophores in addition to 2‑AA and 2‑AB that are widely used in glycan analysis.
Note: Labeling of glycans with 2‑AB is covered under US Patent No. 5,747,347 and its foreign equivalents.
References
To continue reading please sign in or create an account.
Don't Have An Account?