Chromatin Immunoprecipitation (ChIP) Assay
What is a ChIP Assay?
Chromatin immunoprecipitation (ChIP) assays are used to evaluate transcription factor-DNA interactions and are critical for advancing gene expression regulation and epigenetic modifications studies. ChIP can detect and relatively quantify specific protein-DNA and protein-protein interactions in vivo at a single locus or multiple loci. ChIP involves chemically cross-linking proteins to DNA sequences, which is followed by immunoprecipitation of the cross-linked complexes (figure 1), and analysis of the resultant DNA by endpoint or quantitative polymerase chain reaction (qPCR) (figures 2-4), microarrays (ChIP-chip), or next-generation sequencing (ChIP-seq) (figures 5 and 6).
Chromatin immunoprecipitation (ChIP) assay workflow

Figure 1.ChIP procedure workflow
Song et al., 2015
Protein and associated chromatin in living cells or tissues are cross-linked using formaldehyde. The cross-linked DNA–protein complexes (chromatin-protein) are then sheared into ∼500 bp DNA fragments using either enzymatic digestion or physical shearing by sonication. The DNA-protein complexes are then immunoprecipitated by an appropriate protein-specific antibody. After the cross-links are reversed, the associated DNA fragments are eluted, which is followed by immunoprecipitation of the cross-linked complexes and analysis of the resultant DNA by endpoint or quantitative polymerase chain reaction (qPCR), microarrays (ChIP-chip), or next-generation sequencing (ChIP-seq).
ChIP assays can be used for the following studies:
- DNA sequences occupied by multiple specific protein targets
- Binding sites and distribution of a particular protein, such as a transcription factors, transcription co-factors, DNA replication factors and DNA repair proteins throughout the entire genome, under specified cellular conditions
- Gene transcription and polymerase activity
- Complex DNA/protein interactions underlying disease phenotypes
- Modifications to proteins, such as histones, that may influence chromatin structure and gene expression
- Nucleosome architecture and regulation of chromosomal maintenance
ChIP enrichment of DNA sequences from cells

Figure 2.ChIP with cell line samples.
Upper panel: Successful ChIP enrichment of DNA sequences associated with the heterochromatin protein 1d gamma (HP1gamma), an important marker of gene repression. Data collected using ChIPAb+™ Hp1gamma antibody/primer set, mouse IgG (non-specific control) and Magna ChIP™ G kit (Product No. 17-10085).
Lower panel: High-throughput (96-well plate) ChIP analysis of multiple protein targets to query gene loci under various conditions using antibody panels and Magna ChIP™ HT96 Kit (Product No. 17-10077).
Effective ChIP assays with tissue samples

Figure 3.ChIP with tissue samples.
Upper panel: Cryosection tissue and isolate region specific tissue sample with provided 1 mm microdissection punch. An example of region-specific tissue isolation is shown in the image. A 300 µm of coronal mouse brain section with region-specific microdissection of the hippocampus (A) and cerebral cortex (B). The isolated tissue was placed above the dissected region (a: hippocampus, b: cortex). The purified tissue is then dispersed with Tissue Stabilizing Solution followed by formaldehyde treatment. Formaldehyde cross-links proteins to DNA to ensure co-precipitation.
Lower panel: Chromatin from various cell lines was subjected to immunoprecipitation with the indicated antibodies using the Magna ChIP™ G Tissue kit (Product No. 17-20000). The IgG used for relative comparison was either Rabbit Purified IgG (Product No. PP64B) or Mouse Purified IgG (Product No. 12- 371B), depending upon the ChIP antibody. Quantitative PCR data is presented as fold relative enrichment to IgG from independent experiments or as % input. For a biological negative control, qPCR was assessed with primers upstream of the Dhfr gene (UpSt (-)). The antibodies and primers used were as follows: Anti-RNA Polymerase II clone CTD4H8 (Product No. 05-623B): 1 µg of mouse monoclonal affinity-purified antibody immunoprecipitated with chromatin of various mouse tissue and qPCR assayed with primers specific for mouse GAPDH promoter and Anti-SP1 (Product No. 07-645): 1 µg of mouse monoclonal affinity-purified antibody immunoprecipitated from chromatin of various mouse tissue and qPCR assayed with primers specific for the mouse Dhfr promoter.
Higher fold enrichment using low amounts of chromatin or shorter ChIP assay procedure time

Figure 4.Our kits produce higher fold enrichment using low amounts of chromatin or shorter procedure than other supplier's kits.
Upper panel: 10,000 cell equivalents of sonicated chromatin prepared from HeLa cells were chromatin immunoprecipitated using 1 µg of purified IgG (Rabbit IgG, Product No. 12-370) or specific antibodies (H3K4Me3, Product No. 17-614; Phospho-CREB, Product No. 17-10131). Each assay used either the reagents and protocol provided with the Magna ChIP™ HiSens Kit (Product No. 17-10460) or those from the low-input ChIP kit from Supplier A or Supplier D. Immunoprecipitation of antibody-associated DNA fragments was verified by qPCR using positive control primers (GAPDH promoter primers for H3K4me3 and c-Fos promoter primers for pCREB) and negative control primers (human ß-globin promoter primers). Results reflect analysis of 2 µL out of 50 µL total DNA per qPCR reaction.
Lower panel: Complete ChIP reaction in 6 hours in flexible strip well format for Imprint® Chromatin Immunoprecipitation Kit (Product No. ChIP1) Comparison of time required for protocol completion from fixation through purification using different Chromatin Immunoprecipitation kits. Each ChIP experiment was performed using each manufacturers recommended protocol and total required times were compared. HeLa cells were counted, fixed, and immunoprecipitated according to the Imprint CHP1 bulletin with optional High Sensitivity method. ChIP assays were performed using Anti-H3K9ac (H9286) and kit provided antibodies against human RNA polymerase II and non-specific mouse IgG. To evaluate ChIP enrichment, SYBR qPCR was conducted targeting the GAPDH promoter region (highly expressed housekeeping gene). Percent Input describes the amount of DNA with and without antibody selection. For these antibodies, apparent enrichment increased with fewer cells per reaction from diminished non-specific pull-down.
ChIP DNAs analyzed by next-generation sequencing (ChIP-seq)


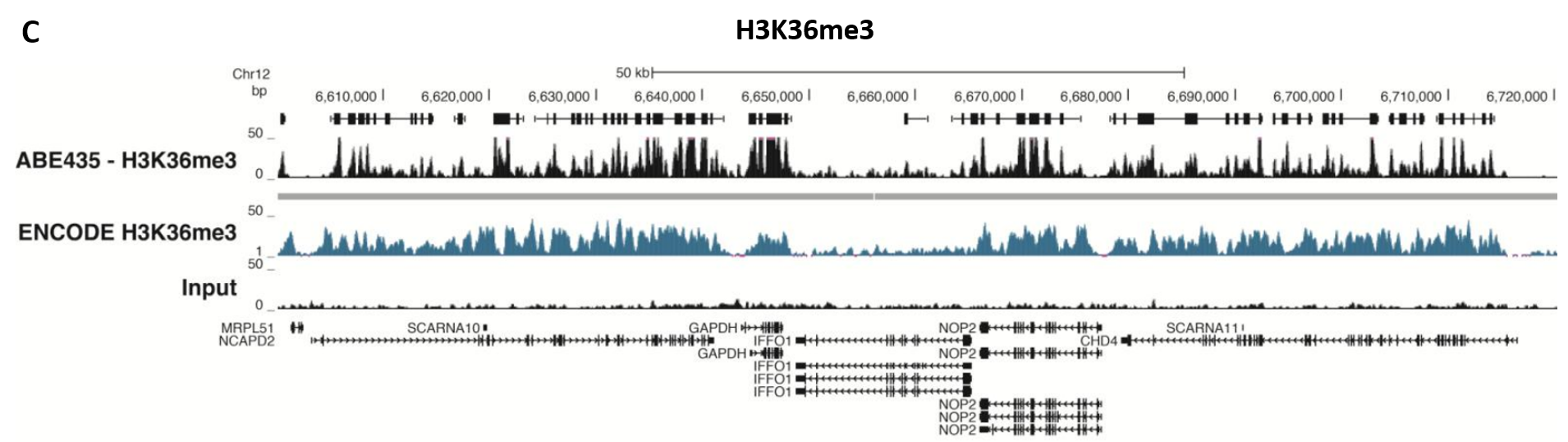
Figure 5.ChIP-Seq Analysis
Chromatin immunoprecipitation was performed using the Magna ChIP™ HiSens kit (Product No. 17-10460), 2 µL, 2 µg, or 5 µg of the anti-H3K4me2 antibody (Product No. 04-790, 05-1338 or ABE250), or 1 or 3 µL or 4 µg anti-H3K4me3 antibody (Product No. 05-745R, 07-473 or 05-1339), or 2 µg anti-H3K36me3 antibody (Product No. ABE435), 20 µL Protein A/G magnetic beads, and 1e6 or 4e6 or 5e6 cross-linked HeLa cell chromatin followed by DNA purification. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on an Illumina HiSeq™ instrument. An excess of twelve million reads from FastQ files were mapped using Bowtie (http://bowtiebio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files. The highest 25% of peaks identified in the data showed 90-99% overlap with peaks identified in the ENCODE H3K4me2 or H3K4me3 or H3K36me3 BROAD Histone tracks for HeLa S3. Data in the region of the transcriptionally active housekeeping gene TUBA1A (tubulin, alpha 1A), HOXB, ACTB or GAPDH is shown.
ChIP assay and next-generation sequencing using low amounts of DNA

Figure 6.Effective ChIP and reliable next-generation sequencing library construction from limited amounts of DNA.
Next-generation sequencing libraries were constructed from Sp1 ChIP DNA using either 10 ng (upper panel) or 1 ng of quantitated immunoprecipitated DNA (middle panel). A reference library was created using 10 ng of purified DNA from the input chromatin. Samples were prepared using the Magna ChIP-Seq Kit (Product No. 17-1010) and a ChIPab+ Sp1 antibody/primer set (Product No. 17-601). The resulting libraries were sequenced on an Illumina Genome Analyzer then aligned and mapped to the human hg18 reference genome. Shown above is the resulting peak analysis (derived using QuEST) of the Sp1 locus from confidently mapped reads browsed with DNAnexus™ software. Replicate sequencing runs from 10 ng and 1 ng libraries are compared, demonstrating overlapping peak identification at the promoter of the Sp1 gene. Triangles reflect high probability Sp1 binding events associated primarily near transcriptional start sites in this genomic interval.
ChIP Experiment Guides, FAQ, Troubleshooting Tips and Supplementary Protocols
ChIP can be challenging, even for the most experienced researcher. It is a multi-step technique (Figure 1) that requires high-quality chromatin, robust antibodies, optimized reagents, and protocols to produce reliable and reproducible results. These are typically achieved by either using commercially prepared products and protocols, or undertaking multiple validation experiments. Regardless of which strategy you prefer, below are a comprehensive ChIP experiment guides for you to tackle some of the problems commonly encountered in ChIP, from sample preparation to downstream DNA analysis.
- Guides to cell numbers for ChIP and endpoint analysis
- Agarose beads vs. magnetic beads
- Crosslinking proteins to DNA and cell lysis
- Immunoprecipitation, washing and elution
- ChIP-qPCR and data analysis (% input and fold enrichment)
- Guide to peak calling for ChIP-Seq
- FAQs (Antibodies, fusion tag, cross-link and beads, chromatin fragmentation and data analysis)
- Troubleshooting tips (high background, low DNA recovery, No DNA amplification, pull down only large DNAs, un-specific DNA precipitate)
- Supplementary protocols
Which ChIP kit is right for you?
Standard ChIP kits
Speciality ChIP kits
ChIP Accessory products
To continue reading please sign in or create an account.
Don't Have An Account?