03-310
In Vivo SHAPE Reagent for Live Cell RNA Structure Analysis
permits the analysis of RNA structure in living cells
Synonym(s):
SHAPE reagent
Sign Into View Organizational & Contract Pricing
All Photos(2)
About This Item
UNSPSC Code:
12161503
eCl@ss:
32161000
NACRES:
NA.32
Recommended Products
Quality Level
form
liquid
manufacturer/tradename
Upstate®
concentration
2 M
technique(s)
activity assay: suitable (reverse transcriptase)
shipped in
dry ice
General description
Features & Advantages
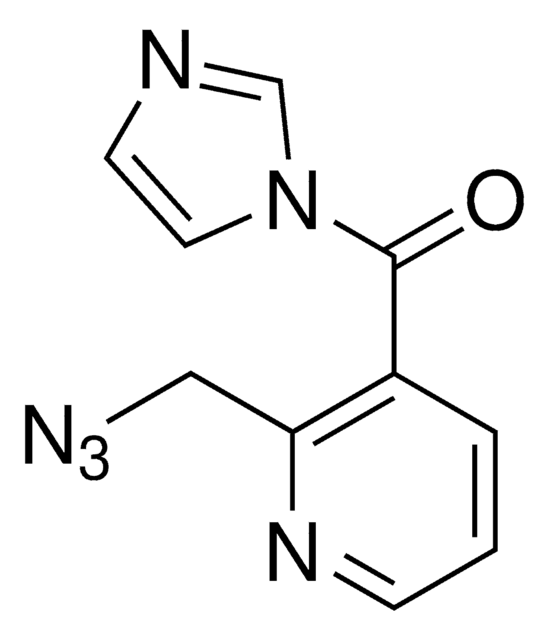
In Vivo SHAPE Reagent is used for the determination of RNA structure. This reagent contains a highly pure form of 2-methylnicotinic acid imidazolide (NAI). This compound forms 2’-O-adducts on single stranded regions of RNA that can be detected by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE).
In contrast to other reagents used for the SHAPE method, the NAI–based In Vivo SHAPE Reagent is rapidly absorbed into cells and is formulated to have low levels of toxicity making it ideal for use with live cells. Unlike other reagents used for SHAPE analysis, In Vivo SHAPE Reagent reacts in an unbiased fashion with all four RNA nucleotides. This combination of low toxicity, rapid uptake and lack of nucleotide bias enables high resolution RNA structure analysis in a variety of living systems.
- Ready-to-use RNase-free solution
- Low toxicity allows use with live cells
- High cell-permeability for rapid modification of RNA
- Reacts with all RNA nucleotides without bias for high resolution mapping
In Vivo SHAPE Reagent is used for the determination of RNA structure. This reagent contains a highly pure form of 2-methylnicotinic acid imidazolide (NAI). This compound forms 2’-O-adducts on single stranded regions of RNA that can be detected by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE).
In contrast to other reagents used for the SHAPE method, the NAI–based In Vivo SHAPE Reagent is rapidly absorbed into cells and is formulated to have low levels of toxicity making it ideal for use with live cells. Unlike other reagents used for SHAPE analysis, In Vivo SHAPE Reagent reacts in an unbiased fashion with all four RNA nucleotides. This combination of low toxicity, rapid uptake and lack of nucleotide bias enables high resolution RNA structure analysis in a variety of living systems.
Folding into higher-order structures allows RNAs to function inside living cells and form dynamic complexes with effector proteins. Predicting these 3D structures is challenging and therefore a number of probing techniques have been developed to study RNA folding.
Compounds such as 2-methylnicotinic acid imidazolide (NAI) contained the In Vivo SHAPE Reagent result in the formation of 2-O-adducts. These adducts form rapidly on single stranded unpaired RNA and much more slowly on based paired (double stranded) RNA. Modified RNA can then be analyzed by primer extension. Modified nucleotides are not extended by RNA dependent DNA polymerases thus allowing sites of modification to be detected.
Compounds such as 2-methylnicotinic acid imidazolide (NAI) contained the In Vivo SHAPE Reagent result in the formation of 2-O-adducts. These adducts form rapidly on single stranded unpaired RNA and much more slowly on based paired (double stranded) RNA. Modified RNA can then be analyzed by primer extension. Modified nucleotides are not extended by RNA dependent DNA polymerases thus allowing sites of modification to be detected.
Components
2-methylnicotinic acid imidazolide (NAI) in DMSO
Quality
Purity of NAI compound is tested by HPLC.
Physical form
0.5 mL vial containing 2M NAI (>97% pure) in DMSO solution
2M NAI in DMSO
Storage and Stability
Store NAI at -20°C in a non-frost-free freezer protected from light. With proper storage (protect from light and avoid excessive freeze-thaw cycles) this product is stable for up to 6 months from the date of receipt.
Legal Information
UPSTATE is a registered trademark of Merck KGaA, Darmstadt, Germany
Disclaimer
Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
related product
Product No.
Description
Pricing
Storage Class Code
10 - Combustible liquids
WGK
WGK 3
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Kevin A Wilkinson et al.
Nature protocols, 1(3), 1610-1616 (2007-04-05)
Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) interrogates local backbone flexibility in RNA at single-nucleotide resolution under diverse solution environments. Flexible RNA nucleotides preferentially sample local conformations that enhance the nucleophilic reactivity of 2'-hydroxyl groups toward electrophiles, such as
Robert C Spitale et al.
Nature chemical biology, 9(1), 18-20 (2012-11-28)
RNA structure has important roles in practically every facet of gene regulation, but the paucity of in vivo structural probes limits current understanding. Here we design, synthesize and demonstrate two new chemical probes that enable selective 2'-hydroxyl acylation analyzed by
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service