17-648
ChIPAb+ Dimethyl-Histone H3 (Lys9) - ChIP Validated Antibody and Primer Set
serum, from rabbit
Synonym(s):
H3K9me2, Histone H3 (di methyl K9), Histone H3K9me2
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
UNSPSC Code:
12352203
eCl@ss:
32160702
NACRES:
NA.52
Recommended Products
biological source
rabbit
Quality Level
antibody form
serum
clone
polyclonal
species reactivity
human, mouse
species reactivity (predicted by homology)
mammals
manufacturer/tradename
ChIPAb+
Upstate®
technique(s)
ChIP: suitable
immunoprecipitation (IP): suitable
western blot: suitable
NCBI accession no.
UniProt accession no.
shipped in
dry ice
Gene Information
human ... H3F3B(3021)
General description
All ChIPAb+ antibodies are individually validated for chromatin precipitation, every lot, every time. Each ChIPAb+ antibody set includes control primers (tested every lot by qPCR) to biologically validate your IP results in a locus-specific context. The qPCR protocol and primer sequences are provided, allowing researchers to validate ChIP protocols when using our antibody in their chromatin context. Each set also includes a negative control antibody to ensure specificity of the ChIP reaction.
The ChIPAb+ Dimethyl-Histone H3 (Lys9) set includes the anti-dimethyl-histone H3 (Lys9) antibody, a negative control antibody (normal rabbit serum), and qPCR primers which amplify a 110 bp region within the promoter of the human β-globin gene. The dimethyl-histone H3 (Lys9) and negative control antibodies are supplied in a scalable "per ChIP" reaction size and can be used to functionally validate the precipitation of dimethyl-histone H3 (Lys9) associated chromatin.
The ChIPAb+ Dimethyl-Histone H3 (Lys9) set includes the anti-dimethyl-histone H3 (Lys9) antibody, a negative control antibody (normal rabbit serum), and qPCR primers which amplify a 110 bp region within the promoter of the human β-globin gene. The dimethyl-histone H3 (Lys9) and negative control antibodies are supplied in a scalable "per ChIP" reaction size and can be used to functionally validate the precipitation of dimethyl-histone H3 (Lys9) associated chromatin.
The methylation of histones can occur on two different residues: arginine or lysine. Histone methylation can be associated with transcriptional activation or repression, depending on the methylated residue. Lysine 9 of histone H3 can be mono-, di- or trimethylated by different histone methyltransferases (HMTs) such as SuvH39H1 or G9a. This methylated lysine can be demethylated by histone demethylases as JMJD1A, LSD1 or JMJD2C. Methylation of this residue is mainly associated with transcriptional repression.
Specificity
Dimethyl-Histone H3 (Lys9)
Immunogen
Epitope: Dimethyl Lys9
The dimethyl-histone H3 (Lys9) rabbit serum is made against a KLH-conjugated, branched synthetic peptide containing the sequence ..Rme2KSTG.., in which me2K corresponds to dimethyl-lysine at residue 9 of human histone H3
Application
Chromatin Immunoprecipitation:
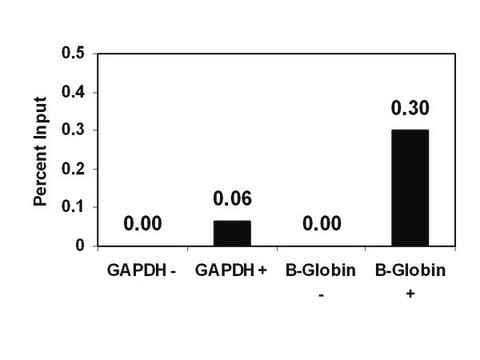
Sonicated chromatin prepared from untreated HeLa cells (1 X 106 cell equivalents) was subjected to chromatin Immunoprecipitation using 4 μL of either a normal rabbit antiserum or Antidimethyl-Histone H3 (Lys9) serum and the Magna ChIP A (Cat. #17-610) Kit (Please see figures). Successful Immunoprecipitation of dimethyl-histone H3 (Lys9) associated DNA fragments was verified by qPCR using β-globin ChIP Primers flanking the human β-globin promoter or primers amplifying the promoter of human GAPDH, which is transcriptionally inactive in HeLa cells. Percent Input relative to standard curves for each qPCR primer set are shown.
Please refer to the EZ-Magna A ChIP (Cat. # 17-408) or EZ-ChIP (Cat. # 17-371) protocol for experimental details.
Western blot analysis and peptide inhibition:
HeLa Acid extract were resolved by electrophoresis, transferred to nitrocellulose and probed with anti-dimethyl-Histone H3 (Lys9) (1:500, Lane 1) or preincubated with 0.4 μM Histone H3 peptide with following modifications:
Lane 2: Linear non-modified
Lane 3: Branched non-modified
Lane 4: Branched trimethyl
Lane 5: Linear trimethyl
Lane 6: Branched dimethyl
Lane 7: Linear dimethyl
Lane 8: Branched monomethyl
Lane 9: Linear monomethyl
Proteins were visualized using a goat anti–rabbit secondary antibody conjugated to HRP and a chemiluminescence detection system.
Sonicated chromatin prepared from untreated HeLa cells (1 X 106 cell equivalents) was subjected to chromatin Immunoprecipitation using 4 μL of either a normal rabbit antiserum or Antidimethyl-Histone H3 (Lys9) serum and the Magna ChIP A (Cat. #17-610) Kit (Please see figures). Successful Immunoprecipitation of dimethyl-histone H3 (Lys9) associated DNA fragments was verified by qPCR using β-globin ChIP Primers flanking the human β-globin promoter or primers amplifying the promoter of human GAPDH, which is transcriptionally inactive in HeLa cells. Percent Input relative to standard curves for each qPCR primer set are shown.
Please refer to the EZ-Magna A ChIP (Cat. # 17-408) or EZ-ChIP (Cat. # 17-371) protocol for experimental details.
Western blot analysis and peptide inhibition:
HeLa Acid extract were resolved by electrophoresis, transferred to nitrocellulose and probed with anti-dimethyl-Histone H3 (Lys9) (1:500, Lane 1) or preincubated with 0.4 μM Histone H3 peptide with following modifications:
Lane 2: Linear non-modified
Lane 3: Branched non-modified
Lane 4: Branched trimethyl
Lane 5: Linear trimethyl
Lane 6: Branched dimethyl
Lane 7: Linear dimethyl
Lane 8: Branched monomethyl
Lane 9: Linear monomethyl
Proteins were visualized using a goat anti–rabbit secondary antibody conjugated to HRP and a chemiluminescence detection system.
Dimethyl-Histone H3 (Lys9) ChIP validated antibody & primer set including the ChIP-grade antibody & the specific control PCR primers used for chromatin immunoprecipitation of H3K9Me2.
Research Category
Epigenetics & Nuclear Function
Epigenetics & Nuclear Function
Research Sub Category
Chromatin Biology
Chromatin Biology
Packaging
25 assays per kit, ~4μL per chromatin immunoprecipitation
Components
Anti-Dimethyl-Histone H3 (Lys9) (rabbit serum), 1 vial
Negative ChIP Control serum, 1 vial
ChIP Primers β-globin , 1 vial
Negative ChIP Control serum, 1 vial
ChIP Primers β-globin , 1 vial
Quality
Chromatin Immunoprecipitation:
Sonicated chromatin prepared from untreated HeLa cells (1 X 106 cell equivalents) was subjected to chromatin immunoprecipitation using 4 μL of either a normal rabbit antiserum or 4 μL Anti-Dimethyl-Histone H3 (Lys9) serum and the Magna ChIP A (Cat. #17-610) Kit.
Successful immunoprecipitation of dimethyl histone H3 (Lys9) associated DNA fragments was verified by qPCR using control ChIP Primers flanking the β-globin human promoter (Please see figures).
Sonicated chromatin prepared from untreated HeLa cells (1 X 106 cell equivalents) was subjected to chromatin immunoprecipitation using 4 μL of either a normal rabbit antiserum or 4 μL Anti-Dimethyl-Histone H3 (Lys9) serum and the Magna ChIP A (Cat. #17-610) Kit.
Successful immunoprecipitation of dimethyl histone H3 (Lys9) associated DNA fragments was verified by qPCR using control ChIP Primers flanking the β-globin human promoter (Please see figures).
Target description
17 kDa
Physical form
Dimethyl-Histone H3 (Lys9) (rabbit polyclonal serum). One vial containing 100 μL of antiserum containing 0.05% sodium azide.
Normal Rabbit Serum. One vial containing 100 uL antiserum containing 0.05% sodium azide.
ChIP Primers, β-Globin. One vial containing 75 μL of 5 μM of each primer specific for human β-globin.
FOR: AGG ACA GGT ACG GCT GTC ATC
REV: TTT ATG CCC AGC CCT GGC TC
Normal Rabbit Serum. One vial containing 100 uL antiserum containing 0.05% sodium azide.
ChIP Primers, β-Globin. One vial containing 75 μL of 5 μM of each primer specific for human β-globin.
FOR: AGG ACA GGT ACG GCT GTC ATC
REV: TTT ATG CCC AGC CCT GGC TC
Storage and Stability
Stable for 1 year at -20°C from date of receipt
Analysis Note
Control
Included negative control antibody normal rabbit serum and control primers specific for human β-globin promoter.
Included negative control antibody normal rabbit serum and control primers specific for human β-globin promoter.
Legal Information
UPSTATE is a registered trademark of Merck KGaA, Darmstadt, Germany
Disclaimer
Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
Storage Class Code
10 - Combustible liquids
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Haobin Chen et al.
Carcinogenesis, 31(12), 2136-2144 (2010-10-01)
Epigenetic silencing of tumor suppressor genes commonly occurs in human cancers via increasing DNA methylation and repressive histone modifications at gene promoters. However, little is known about how pathogenic environmental factors contribute to cancer development by affecting epigenetic regulatory mechanisms.
Youhua Tan et al.
Biochemical and biophysical research communications, 483(1), 456-462 (2016-12-23)
Tumor-repopulating cells (TRCs) are a tumorigenic sub-population of cancer cells that drives tumorigenesis. We have recently reported that soft fibrin matrices maintain TRC growth by promoting histone 3 lysine 9 (H3K9) demethylation and Sox2 expression and that Cdc42 expression influences
Valérie Grandjean et al.
Development (Cambridge, England), 136(21), 3647-3655 (2009-10-13)
The size of the mammalian body is determined by genetic and environmental factors differentially modulating pre- and postnatal growth. We now report a control of growth acting in the mouse from the first cleavages to the postnatal stages. It was
Marianna Rodova et al.
Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research, 26(8), 1974-1986 (2011-04-01)
The development of disease-modifying pharmacologic therapy for osteoarthritis (OA) currently faces major obstacles largely because the regulatory mechanisms for the function of adult articular chondrocytes remain unclear. We previously demonstrated that lack of Nfat1, one of the nuclear factor of
Jia-Yi Yao et al.
The Journal of biological chemistry, 285(24), 18828-18837 (2010-04-13)
Stable epigenetic silencing of p16(INK4a) is a common event in hepatocellular carcinoma (HCC) cells, which is associated with abnormal cell proliferation and liberation from cell cycle arrest. Understanding the early epigenetic events in silencing p16(INK4a) expression may illuminate a prognostic
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service