Exploring the Future of Drug Discovery with DNA Encoded Libraries (DELs)
INTRODUCTION
In the quest to discover novel therapeutic agents and revolutionize drug development, scientists have continually sought innovative approaches to accelerate the identification of potential drug candidates. One ground-breaking solution that has emerged in recent years is the utilization of DNA-encoded libraries (DELs). DELs stand as a versatile and potent technology foundation for uncovering small molecule binders for biologically and pharmaceutically significant protein targets.
DELs encompass groups of compounds, each linked to unique DNA markers that function as amplifiable identification codes. With the progress made in DNA-compatible chemical reactions, selection techniques, next-generation sequencing, and data interpretation, DEL technology enables the creation and examination of libraries with unparalleled dimensions.
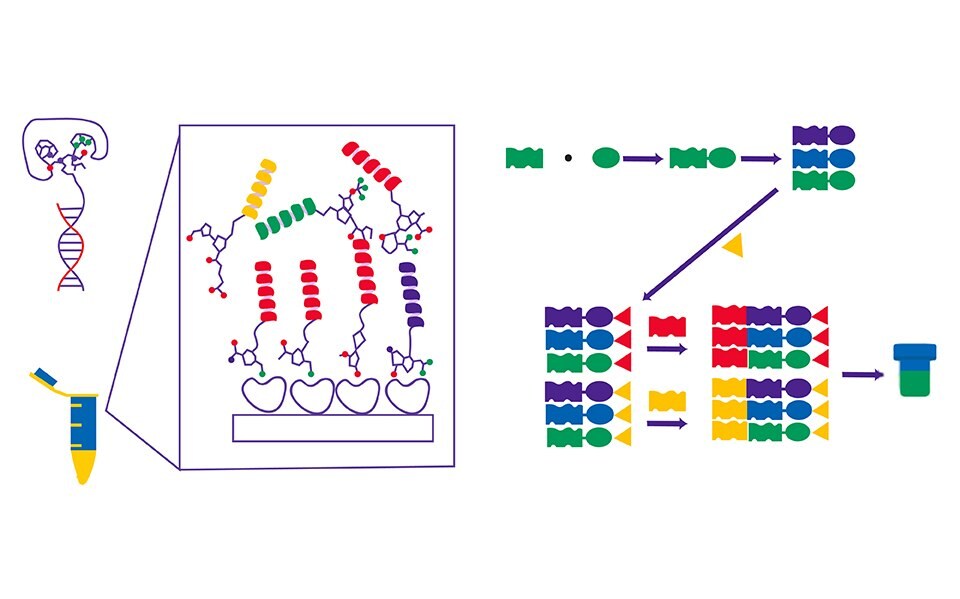
Figure 1.DNA-encoded library and its mode of operation.
The main challenges addressed by DEL technology
Traditional methods of drug discovery are often time-consuming, costly, and inefficient. They rely on screening vast collections of chemical compounds to identify molecules with the desired therapeutic properties. This approach not only demands enormous resources but also limits the scope of the search to a finite chemical space. Additionally, the identification and optimization of lead compounds frequently involve extensive hit-to-lead optimization, further elongating the drug development timeline. Another challenge is the high cost of synthesizing and screening chemical compounds with many promising molecules often being overlooked due to budget constraints.
The technology
DNA encoded libraries offer a paradigm shift in drug discovery by leveraging the power of molecular biology and high-throughput screening. This technology consists of four key elements or features:
- DNA Tagging: Each chemical compound in the library is tagged with a unique DNA sequence, creating a barcode-like identifier for the molecule.
- Massive Library Size: DELs can contain billions to trillions of compounds, greatly expanding the chemical space that can be explored.
- High-Throughput Screening: By utilizing DNA tags, researchers can rapidly and simultaneously screen vast libraries to identify compounds binding to a specific target.
- Data-Driven Approach: The technology enables the generation of large-scale data sets, allowing for data analysis and machine learning approaches to identify potential leads.

- All compounds are in a single vial
- Each attached to a unique DNA barcode

- Target immobilized on a bead
- Screen library all at once
- Wash away compounds that didn’t bind

- Isolate hits
- Amplify DNA with PCR
- Sequence DNA code

- Use code to determine structure of hit
Figure 2. DNA-encoded library main protocol stages (or steps).
Advantages of using DEL technology
DELs provide access to an unparalleled diversity of chemical compounds, increasing the chances of discovering unique drug candidates. In addition, high-throughput screening with DNA tags significantly accelerates the drug discovery process, reducing development timelines, while enabling efficient resource utilization, therefore, making drug discovery more cost-effective. In conclusion, DEL technology opens doors to innovative target-based and phenotypic drug discovery approaches.
Which DNA-encoded library is right for my research? |
|---|
Looking for extended DEL data to use in Artificial Intelligence/Machine Learning
DEL Data for 1 analysis of DNA-Encoded Library information - DELDATA
Introducing our state-of-the-art DNA Encoded Library (DEL) Enhanced Data Bundle, an innovative solution that introduces unmatched effectiveness and creativity to the realm of pharmaceutical research. DEL Data merges the capabilities of DNA labelling with sophisticated analysis of successful hits, enabling scientists to expedite their drug development endeavours and unveil novel therapeutic prospects. This DNA labelling method enables simultaneous screening of countless, even astronomical amounts of compounds, vastly boosting the speed and effectiveness of the screening process. The Enhanced Data Bundle elevates this technology to the next level by offering thorough and intricate insights into the hits acquired during the screening of the DNA-encoded library. It provides researchers with an exhaustive overview of the chemical structures and binding affinities of the successful compounds, giving them the authority to make informed decisions when selecting the most promising candidates for further examination.
REFERENCES
To continue reading please sign in or create an account.
Don't Have An Account?