Single-Cell Resolution Discovery
Pooled CRISPR Screening with 10x Genomics Compatibility
CRISPR lentiviral products are powerful research tools for the discovery of novel genes and cellular pathways essential to understanding human health and disease. As a world leader in the production of lentiviral libraries, we design and build custom pools to any specifications. With this capability, we have partnered with 10x Genomics to bring innovative pooled screening products to enable discovery at single-cell resolution
CRISPR-based functional genomics screening provides a broad overview of the genetic contributions to a measurable phenotype. Bulk screening, however, only provides limited functional information beyond the selectable phenotype. At the other end of the spectrum, single-cell analysis provides high content information centered around genome-wide transcriptome profiling for candidate genes. We offer optimized lentiviral vectors compatible with the Chromium Next GEM Single Cell Gene Expression assays, utilizing Feature Barcode technology from 10x Genomics.
To speak to a representative for more information or requests a quote:
Getting Started with 10x Single Cell Compatible Products: |
|---|
Recommendations
- Consider doing a pilot study using our optimization kit and/or individual clones
- For small pool screening, we recommend 100-200 cells for each targeting guide and
500-1000 cells that contain a non-targeting guide. - Use 2-5 gRNAs per gene target
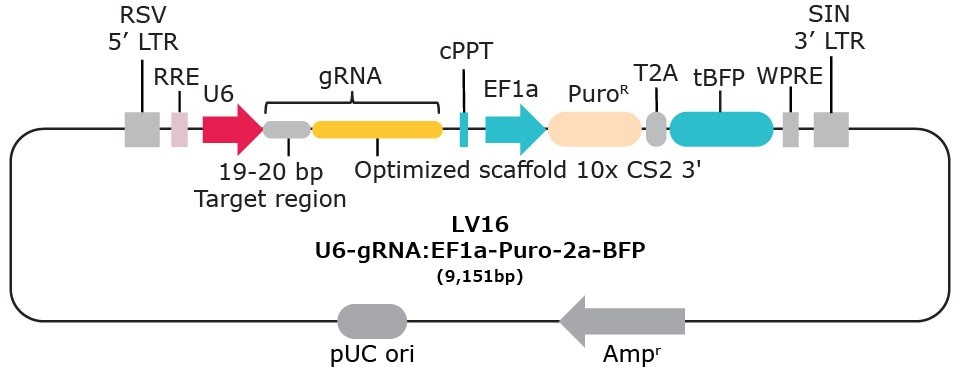
Vector options with 10x single cell compatibility:





Capture Sequence Integration

Figure 1.CRISPRi Feature Barcode technology Optimization Kit
This kit includes pooled virus representing each of the 2 possible capture sequences and 2 position combinations (capture sequences 1 or 2, in the stem-loop or 3′ position). Present within each pool are 2 CRISPRi sgRNAs, 1 targeting RAB1A and 1 non-targeting control sgRNA. Each pool is provided ready to use at 1x10^6 viral titer, sufficient enough to cover ~2x10^6 cells at an MOI of 0.5. The purpose of the optimization kit is to set up a pilot screen to determine the best capture sequence and position configuration before a custom pool is ordered.
10x Compatible products for Feature Barcode technology (gRNA)

Figure 2.We recommend creating custom pools so that when running the 10x Single Cell CRISPR assay, 100-200 cells are recovered for each targeting guide and 500-1000 cells are recovered that contain a non-targeting guide. For example, a user that wishes to target 10,000 cells recovered, we recommend that 10% of the guide pool is made up of non-targeting guide(s), while the remaining 90% is targeting guides. For the targeting guides in this example we would recommend that each targeting guide represent >=2% of the overall guide pool. The more cells that are run, the smaller the percentage of the pool is needed to be non-targeting guide(s).
Tracking gene perturbations at single cell resolution
Knockdown efficiences: log2 fold change vs p-value

Figure 3.Results from a small test CRISPRi pool showing the knockdown efficiency of each guide on its target gene through fold change in expression versus normal levels. Non-targeting guides as well as those with lower knockdown sit in the bottom right quadrant while those exhibiting stronger and statistically significant repression are located in the top left. Because of cell line variation and gRNA efficiency it’s important to target with multiple gRNAs per gene basis, we recommend 3-5 per gene.
SINGLE CELL ANALYSIS AND WHOLE TRANSCRIPTOME READOUTS

Figure 4. New, higher-resolution analysis with clustering by both CRISPR guide and transcriptomic profile is possible. A375 cells were transduced with a pool of approximately 100 guides in 10x Genomics-compatible vectors. Each dot represents a single cell (A) Clustering based only on gene expression profile. (B) Clustering based only on CRISPR guide identity. Distinct clustering indicates that the platform can successfully identify unique guides and use that as a basis to explore resulting gene expression changes. (C) Specific cells from (A) representing presence of either a non-targeting guide or a CRISPRi guide targeting ELOF1. (D) Gene expression analysis of ELOF1 in cells from (C) expressing either a non-targeting guide or a guide targeting ELOF1. Expression data shows a significant reduction in the presence of ELOF1 mRNA in cells expressing a CRISPRi guide targeting ELOF1 as proof-of-concept, indicating that it is possible to analyze both transcriptomic profile and gene perturbation simultaneously in single cells.
Other Resources
Protocols for single cell CRISPR screening using lentivirus:
For more information on Feature Barcode technology and Analysis:
For more information see:
References
To continue reading please sign in or create an account.
Don't Have An Account?