Introduction to Proximity Labeling
Understanding how biomolecules interact with each other can lead to fundamental discoveries in biology and have an outstanding impact in understanding the progression of human disease. However, identifying these key interactions can be extremely challenging via traditional methods.
Recently, an unbiased strategy for unveiling these interactions has been developed called Proximity Labeling. Here, a biomolecule of interest is appended to an “antenna” that can catalytically generate reactive species upon stimulation. These reactive species are then able to “tag” nearby biomolecules that can be analyzed downstream to give a list of interacting proteins (or nucleic acids) that can be used to measure their proximity or distance to the antenna (Figure 1).

Figure 1.(Left side) An ‘antenna’ allows proximity labeling of nearby molecules. (Right side) A protein of interest fused to an ‘antenna’ can be used to generate reactive intermediates.
Challenges in Cell Surface Proximity Labeling
The identity of both the antenna and the reactive intermediate have been well explored in recent years with most variations being based on engineered biotin-ligase or peroxidase enzymes. These systems are triggered by the addition of biotin and hydrogen peroxide to provide activated ester and phenoxy radical intermediates, respectively (Figure 1, right). These reactive intermediates possess solution half-life between 1 ms and 1 min; in the crowded environment of the cell, these intermediates rapidly react with biomolecules - limiting free diffusion. However, in the extracellular space, the long solution half-life leads to very large labeling radii and subsequently long lists of identified proteins (Figure 2). This presents a significant issue for practitioners where validation of false positives is frequently required, wasting precious resources.
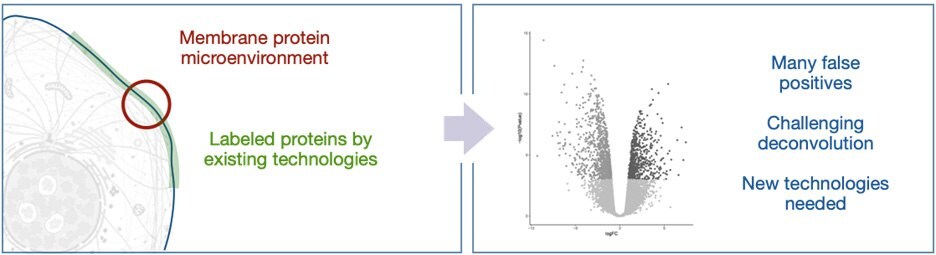
Figure 2.Current proximity labeling in the extracellular space can lead to many false positives.
ATLAS µMap kit for high resolution proximity labeling
Recently, a collaboration between Professor MacMillan’s group at Princeton and Scientists at Merck developed a method to address these issues. They questioned whether it would be possible to provide a higher resolution labeling platform by generating carbene reactive intermediates that display a significantly shorter solution half-life. These carbenes are formed catalytically through the photosensitization of aryl diazirines by iridium catalysts. By tethering these iridium catalysts to an antibody, this method provides short radius mapping of membrane protein microenvironments (Figure 3A).
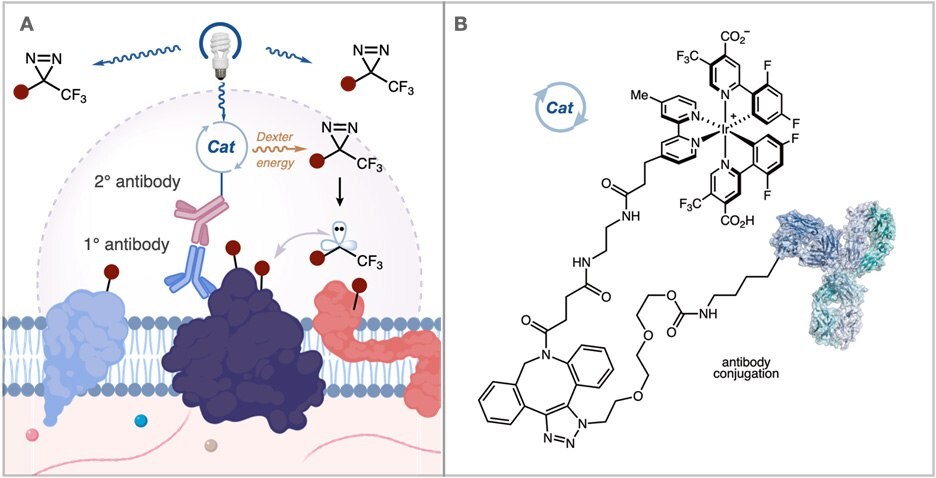
Figure 3.(A) Activation of an iridium catalyst attached to an antibody generates carbene reactive intermediates with a shorter half-life for nearby proximity labeling in the extracellular space. (B) Atlas kits offer a method to attach the iridium catalyst to an antibody of interest.
Advantages
- Facile incorporation onto targeting antibodies
- Short labeling radius
- Benign method of activation (visible light)
- High temporal resolution
- Investigation of any membrane microenvironment with high fidelity
References
To continue reading please sign in or create an account.
Don't Have An Account?