Locked Nucleic Acid & Minor Groove Binder / Eclipse Dark Quencher
Locked Nucleic Acid and Minor Groove Binder / Elipse Dark Quencher (MGB:EDQ) are modifications that enhance the performance of oligonucleotides. They increase stability and improve the specificity and affinity for base-pairing with target sequences. Locked Nucleic Acid enhances primers and probes (when combined with BHQ™) in qPCR as well as the effectiveness of therapeutic modalities like antisense oligonucleotides (ASOs). Meanwhile, MGB:EDQ, with its included quencher, specifically enhances probes in qPCR.
Locked Nucleic Acid
Locked Nucleic Acid is a novel type of nucleic acid analog that contains a 2'-O, 4'-C methylene bridge (Figure 1). This bridge–locked in the 3'-endo conformation–restricts the flexibility of the ribofuranose ring and locks the structure into a rigid bicyclic formation. This confers enhanced assay performance and an increased breadth of applications.

Figure 1.Structure of Locked Nucleic Acid and native-state DNA monomers.
Locked Nucleic Acid in PCR primers, qPCR probes, and other types of oligonucleotides is soluble in water and standard buffers as well as follows Watson-Crick base-pairing rules.1
Benefits of Locked Nucleic Acid
When incorporated into oligonucleotides, Locked Nucleic Acid offers several benefits compared to native-state DNA bases only, including the following:
- Increased thermal stability and hybridization specificity
- More accurate gene quantification and allelic discrimination
- Easier and more flexible designs for problematic target sequences
Increased Thermal Stability and Hybridization Specificity
Incorporating Locked Nucleic Acid into oligonucleotides increases thermal duplex stability2 and improves the specificity of oligonucleotide hybridization to target sequences.3 In the case of qPCR, this reduces background fluorescence from spurious binding, which increases the signal-to-noise ratio. In addition, the improved hybridization of the PCR primer or qPCR probe to its target may increase the melting temperature (Tm) by up to 8 °C per Locked Nucleic Acid monomer substitution in medium salt conditions compared to native-state DNA oligonucleotides4 (Table 1). This increase in hybridization creates a significant broadening in the scope of assay conditions and allows for more successful multiplexing.5
More Accurate Gene Quantification and Allelic Discrimination
The ability of oligonucleotides to discriminate between alleles via SNPs is greatly enhanced by the incorporation of Locked Nucleic Acid bases6-8 (Figure 2). The presence of a single base mismatch has a greater destabilizing effect on the duplex formation between a Locked Nucleic Acid oligonucleotide and its target than with a native-state DNA oligonucleotide. Shorter oligonucleotides incorporating Locked Nucleic Acid bases can be used at the same temperatures as longer native-state DNA oligonucleotides.

Figure 2.Locked Nucleic Acid Dual-Labeled Probes discriminate better than DNA Dual-Labeled Probes in SNP genotyping analysis9. Pink) mutant DNA analysis with Locked Nucleic Acid mutant probe (16mer with 3 Locked Nucleic Acid bases). Green) mutant DNA analysis with native-state DNA mutant probe (25mer). Red) wild-type DNA with native-state DNA mutant probe (25mer). Purple) wild-type DNA with Locked Nucleic Acid mutant Probe (16mer with 3 Locked Nucleic Acid bases).
Easier and More Flexible Designs for Problematic Target Sequences
Due to Locked Nucleic Acid’s improved hybridization characteristics with accompanied increase in Tm, Locked Nucleic Acid oligonucleotides can be synthesized to be shorter, which overcomes certain design limitations that occur with native-state DNA oligonucleotides. Specifically, Locked Nucleic Acid oligonucleotides can be designed to address traditionally problematic target sequences, such as AT- or GC-rich regions. For example, AT-rich native-state DNA oligonucleotides often need to be over 30 bases long (sometimes over 40 bases) to satisfy amplicon design guidelines but may still perform poorly. With Locked Nucleic Acid oligonucleotides, the selective placement of Locked Nucleic Acid bases facilitates the optimal design of highly-specific, shorter oligonucleotides that perform well, even at lengths of only 13 to 20 bases. Also, the design of oligonucleotides for targeting difficult SNPs, such as the relatively stable G:T mismatch, is greatly facilitated by Locked Nucleic Acid.
Additional Benefit
Locked Nucleic Acid PCR primers and qPCR probes are compatible with all real-time thermocyclers and end-point analytical detection instruments. No specialized instruments are necessary.
Applications of Locked Nucleic Acid
Locked Nucleic Acid can be incorporated into all available qPCR detection chemistries, including:
- SYBR® Green Primers
- Dual-Labeled Probes
- Molecular Beacons
- LightCycler® Probes
- Scorpions® Probes
It is useful for the following:
- SNP detection
- Allele discrimination
- Pathogen detection
- Multiplexing
- Viral load quantification
- Gene expression analysis
- Gene copy determination
It can also be used in these sequences:
- Antisense oligonucleotides
- Decoy oligonucleotides
- Capture probes
- Aptamers
- Ribozymes
MGB:EDQ
An MGB:EDQ Probe is a single-stranded oligonucleotide labeled with two different dyes. A reporter dye is located at the 5’ end and Minor Groove Binder / Eclipse Dark Quencher is located at the 3’ end (Figure 1). The quencher molecule inhibits the natural fluorescence emission of the reporter by fluorescence resonance energy transfer (FRET, Figure 2).
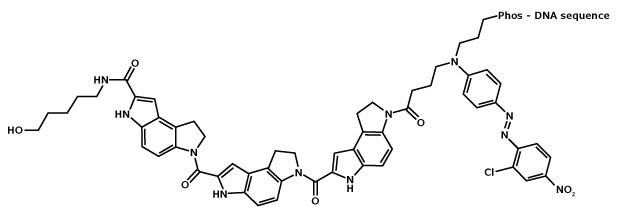
Figure 1.Structure of MGB:EDQ. MGB, or CDPI3, is the tripeptide of dihydropyrroloindole-carboxylate, and its boomerang curvature allows it to align to the shape of and thereby strongly bind to the minor groove of B-form DNA.
1. Probe in solution emits low fluorescence
2. Emission of the fluorescence by hydrolysis
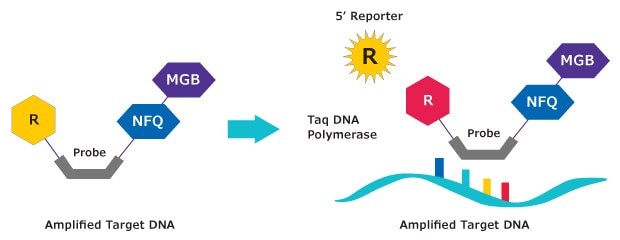
Figure 2.Mechanism of action. MGB:EDQ Probes are hydrolysis probes. The primer is elongated by the polymerase and the probe binds to the specific DNA template. Hydrolysis releases the reporter from the probe/target hybrid, causing an increase in fluorescence. The measured fluorescence signal is directly proportional to the amount of target DNA.
BENEFITS OF MGB
MGB enhances the performance of qPCR in several ways:
- Increased specificity: MGB increases the specificity of the probe-target hybridization, reducing the likelihood of non-specific amplification
- Higher sensitivity: MGB enables the use of shorter probes, which increase the sensitivity of qPCR assays
- Melting temperature adjustment: MGB allows for Tm adjustment of the probe, which can be beneficial for designing qPCR probes with optimal Tm values
- Reduced background signal: MGB contributes to a reduction in background fluorescence, leading to an improved S:N or signal-to-noise ratio
- Enhanced stability: MGB can improve the stability of the probe-target hybrid, reducing the likelihood of probe degradation
- Design flexibility: The use of MGB expands the options for probe design, facilitating the creation of efficient qPCR assays
MGB-enhanced qPCR probes are a valuable tool for achieving reliable and robust qPCR results.
APPLICATIONS OF MGB:EDQ
MGB:EDQ can be incorporated into this qPCR detection chemistry:
- Dual-Labeled Probes (with 6-FAM™, HEX™, and TET™; inquire for others)
It is useful for the following:
- SNP detection via mismatch discrimination
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?