Rapid Determination of Protein Binding Affinity Using Solid Phase Microextraction
Emily R, Barrey
Senior Scientist, R&D and Craig Aurand, Innovation Manager, Advanced Analytical
Reporter US Volume 34.4
Determination of free circulating drug concentration is important in establishing pharmacokinetic activity. In most cases, drug-protein complexes could be formed which affect the active level of circulating drugs in the body. Techniques used for determining drugprotein binding levels consist of ultrafiltration, ultracentrifugation, and microdialysis. Automation can be used in the case of microdialysis, but processing may be greater than 6 hours due to the need to reach equilibrium. In this study, a novel biocompatible microextraction device is evaluated as a rapid means of extracting the non-bound fraction of drugs from plasma. Initial studies demonstrate that drug binding affinities can be determined in less than 60 minutes using this microextraction technique. A model set of protein binding drugs was selected and their free concentrations were compared using the SPME LC (BioSPME) approach and the equilibrium dialysis technique (rapid equilibrium dialysis device-RED device). Drugs with reference binding affinities ranging from 20%-99% were selected for comparison of the sampling devices.
The BioSPME technique enables direct analysis of biological samples without the need for protein precipitation, centrifugation, or digestion. The BioSPME technique allows for isolation of target analytes while minimizing coextraction of sample matrix, allowing for more sensitive and robust analysis. The extraction mechanism for the SPME LC utilizes differential migration that is dependent upon the affinity of the analyte for the phase coating on the fiber to the affinity for the matrix. In the case of the SPME LC fibers, the polymeric binder used to adhere the phase onto the fiber core acts as a shield that prevents large molecular weight molecules (i.e. proteins) from absorbing onto the fiber, thus allowing for only the free fraction (unbound) analyte to be extracted by the fiber coating. Figure 1 depicts a representation of the extraction mechanism for the SPME LC fibers.

Figure 1.Extraction Mechanism for SPME LC Fibers
Experimental
Custom standard mixes containing codeine, diazepam, diclofenac, propranolol, warfarin, and quinidine were prepared. Labeled internal standards were also obtained for all of the compounds except quinidine. Rat plasma stabilized with K2EDTA (BioReclamation, IVT, Hicksville, NY USA) was spiked at 200 ng/mL of binding analytes and allowed to equilibrate for 3 hours at 37 °C prior to extraction studies. Phosphate buffered saline (PBS, pH 7.4) was prepared with the following concentrations: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4. PBS was spiked at 200 ng/mL with the analytes of interest and this solution was used for the SPME extraction studies. Blank PBS was used for the RED device binding studies. Samples were analyzed using an Agilent 1290 Infinity II UPLC with a 6460 QQQ mass spectrometer. The mass spectrometer parameters are listed in Table 1.
SPME LC C18 Preparation
Fibers were conditioned by soaking in methanol for 10 minutes. Fibers were then equilibrated by soaking in water for 10 minutes. The samples were prepared by placing 800 μL of plasma and buffer samples into a 2.0 mL Nunc® 96-well plate. The SPME extraction was conducted by placing the SPME C18 fibers directly into 800 μL of plasma or buffer and agitated for 30 minutes at 500 rpm. Five sample replicates were prepared. Fibers were then transferred from the samples and placed directly into a 600 μL conical 96-well plate that had been prefilled with 300 μL of internal standard desorption solvent (50 ng/mL in acetonitrile). The samples were desorbed in the well plate by agitating for 10 minutes at 500 rpm. The SPME fibers were removed, and the well plate was capped, vortexed, and analyzed directly.
RED Device Preparation
The RED Device samples were prepared by placing 200 μL of plasma into the plasma compartment of the RED device and 350 μL of buffer into the buffer compartment. The RED device was capped with a sealing mat and equilibrated at 37 °C at 250 rpm for 4 hours. After the equilibration step, 50 μL aliquots were taken from each compartment, 50 μL of plasma was added to the buffer compartment aliquot, and 50 μL of the buffer was added to the plasma compartment aliquot. After mixing, 300 μL of ice cold acetonitrile with 50 ng/mL internal standard were added. Samples were vortexed at 1200 rpm for ~ 5 minutes and then centrifuged at 15,000 rpm for 10 minutes. The supernatants were decanted into a glass HPLC vial for analysis.
Results
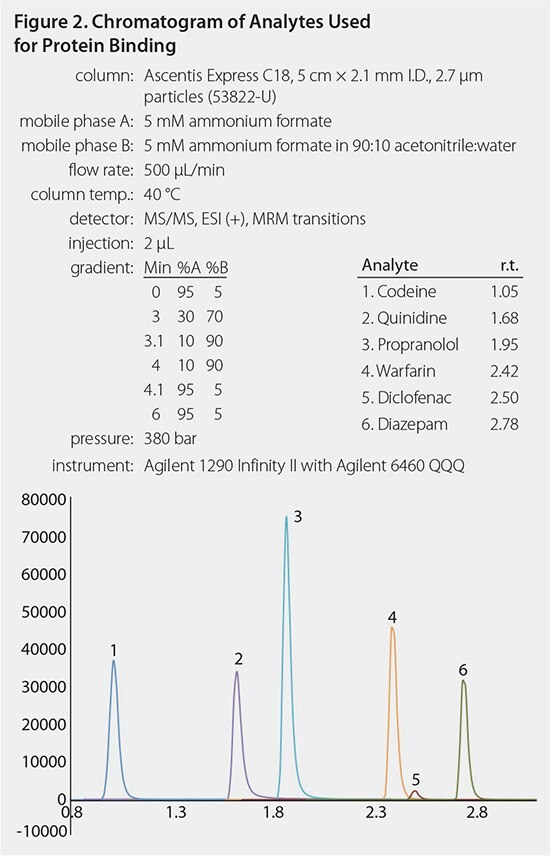
The total SPME processing time took ~ 40 minutes, whereas the total processing time for the RED Device was ~ 5 hours. A typical chromatogram and calibration curve are shown in Figures 2 and 3. Analyte binding concentrations were calculated with the following equations:



Conclusions
In the case of the SPME LC analysis, the free fraction of the analyte is measured in both the reference (PBS) and the plasma sample. This technique simplifies the calculation for determining the protein binding level. The SPME LC approach allows for direct sampling of the plasma sample, eliminating the need for protein precipitation as in the equilibrium dialysis device. The ability to concentrate analytes onto the fiber and desorb them into smaller volumes enables lower detection limits. This approach will be beneficial especially for those analytes that have high binding affinities and low MS sensitivity. This direct sampling approach also minimizes concern associated with matrix interference. Plasma protein precipitation will not remove matrix interferences like phospholipids, whereas the phospholipids will not be extracted onto the SPME fiber.
Protein binding affinities for both the SPME LC and the equilibrium dialysis devices closely matched the referenced range for all analytes. By utilizing the SPME approach, binding affinities correlating to the reference values were obtained in a significantly shorter time span compared to membrane dialysis techniques.
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?