Oligonucleotide FAQ
Oligonucleotides are short DNA or RNA molecules that are used in genetic testing, research, and forensics. Custom oligos are synthesized using phosphoramidite chemistry.
Our customer service representatives and technical service scientists are more than happy to answer any questions that you may have regarding oligonucleotides. However, you might find the answer you need among some of the most frequently asked questions regarding our custom oligo synthesis service.
OLIGO SYNTHESIS
How are oligonucleotides synthesized?
Oligonucleotides are synthesized using phosphoramidite chemistry. Details of our synthesis methods can be found in our DNA Oligonucleotide Synthesis article.
OLIGO LENGTH
What is the longest sequence that I can order?
Standard DNA Oligos are available up to 120 bases. However, Long Oligos—specifications are more limited compared to Standard DNA Oligos—are available up to 180 bases. For details, please review our Long Oligos offering.
What is the shortest sequence that I can order?
Standard DNA Oligos are available as short as 6 bases. If you need less than 6 bases, please contact dnaoligos@sial.com for feasibility assessment / quotation.
CUSTOM MODIFICATIONS
Does my oligonucleotide have a phosphate on the 5' or 3' end?
All oligonucleotides are synthesized with a hydroxyl group on each end. However, phosphate can be added to both ends. Please add this modification to your sequence in either the online ordering configurator or the email ordering template. The code for phosphate is [Phos] and looks like this in the sequence:
[Phos]ACGTACGTACGTACGT[Phos]
What modifications are available?
Standard modifications available by category (fluorescent, attachment, binding, spacer, analog, intercalation, antisense, and phosphorothioate) can be found on our Custom DNA Oligos Modifications page. If you do not see your required modification, there is a good chance that we can still work with it, especially if it is commercially available as a CPG, phosphoramidite, or NHS ester.
If not commercially available, there is a chance that we can manufacture your modification as a raw material (for feasibility, please email dnaoligos@sial.com with a description of your needs; a minimum spend of $25,000 and a lead time of six months often applies).
When designing my sequence, where should I place modifications?
This depends on the requirements of the intended technique and application. For example, with a qPCR probe, a 5' reporter dye and a 3' quencher is considered standard, is most commonly requested, and works well. However, some users may require an internal quencher for a variety reasons, which we might be able to manufacture upon request. For qPCR probes that require shorter sequence lengths due to targets with AT-rich regions, LNA® (Locked Nucleic Acid®) can be added internally. For assistance with modification placement, please contact our technical services team at oligotechserv@sial.com.
Do you offer alternative dyes to those that are protected by intellectual property?
Yes, if you intend to commercialize your oligonucleotides, it is understandable that you want dyes giving you freedom to operate. Please contact our technical services team at oligotechserv@sial.com, and we will determine what alternatives are available for the dyes you want to replace.
OLIGO PURITY
What purification methods are available and what are their guaranteed purities?
Our methods and guaranteed purities, include: desalt (no guarantee), cartridge (no guarantee, but 65-80% full-length sequence is typical), RP-HPLC (Reverse-Phase High Performance Liquid Chromatography, >85% full-length sequence), and PAGE (Polyacrylamide Gel Electrophoresis, >95% full-length sequence). While less-commonly requested, we also offer IE-HPLC (Ion-[specifically, Anion]-Exchange HPLC, please inquire for the guaranteed full-length sequence). Finally, we also offer NGSO (Next-Gen Sequencing Oligos), which are custom adapters purified with proprietary processes, the purposes of which are to drive down cross contamination to the lowest levels possible. Details of our purification methods can be found in our Oligonucleotide Purification article.
Why do the analytical HPLC chromatograms of phosphorothioate oligonucleotides appear broad?
The presence of sulfur creates a stereogenic α-phosphorus, and the resulting diastereomers have subtly shifted elution times during chromatography. Details of the problem can be found in our Chromatography Profile Analysis of Phosphorothioate Oligonucleotides article.
OLIGO QUANTITY DETERMINATION
How do you determine the quantity?
We use the following method to determine oligo quantity: 1) measure A260 values using a UV-Vis spectrophotometer; 2) convert the absorbance value to concentration using the Beer-Lambert Law; 3) convert the concentration to customary units of optical density (OD). Details and example calculations of the method can be found in our Oligonucleotide Quantification article.
If I order the 0.2 µmol scale, will I receive 0.2 µmol of material?
No, because the amount of material delivered is always less than the starting synthesis scale (you can request an exact amount of material as opposed to ordering by starting synthesis scale). This is because the coupling efficiency between each base is <100%. In addition, cleavage, deprotection, and purification decrease the yield further (see the section titled, ‘Yield’ at the bottom our DNA Oligonucleotide Synthesis article for additional details). For minimum yield guarantees, please review our Standard DNA Oligos offering.
OLIGO FORMATS
Why do I not see any material in the vessel (tube / plate well)?
Unless specified differently, all oligonucleotides are provided dry. If you hold the tube / plate in light at the right angle, you might see the material as a thin film or glaze at the bottom of the vessel. Before removing caps / plate mats, we recommend centrifugation just in case the material came loose during transit. Resuspension in water or buffer and an A260 reading should provide confidence that the oligonucleotide is present.
OLIGO SERVICES
Do you anneal oligonucleotides?
Yes, please submit a request to dnaoligos@sial.com for feasibility review / quotation. You may also view our Protocol for Annealing Oligonucleotides, if you wish to do it yourself.
OLIGO MELTING TEMPERATURE
How is the melting temperature (Tm) calculated?
For sequences ≥15 bases, we use the nearest neighbors method. For sequences ≤14 bases, we use a basic method (modification of the Marmur Doty formula). Example calculations for each method are detailed in our Oligonucleotide Melting Temperature article.
Why are the melting temperatures (Tm) on my technical datasheet different than those calculated by OligoArchitect™ Online?
OligoArchitect™ Online is optimized for qPCR whereas the methods used by our internal production system are optimized for PCR. If you have used OligoArchitect™ Online to design your sequences, then use those Tm in your experiments. If you did not use OligoArchitect™ Online to design your sequences, and you intend to use your oligonucleotides in PCR, then use the Tm provided on the technical datasheet.
OLIGO HANDLING
How do I make a 100 µM stock solution?
A shortcut calculation is as follows: take the number of nmol from the tube label / technical datasheet and multiply by 10 to get the resuspension volume in µL (microliters; this calculation is actually unnecessary as the volume needed for resuspension is included on the technical datasheet). Details and example calculations of longform and shortcut calculations can be found in our Oligonucleotide Handling & Stability article.
How do I convert nmol (nanomoles) to µg (micrograms)?
This example use longform dimensional analysis:
- Assume oligonucleotide amount of 30 nmol
- Assume molar mass (molecular weight) of 6766.5 g mol-1
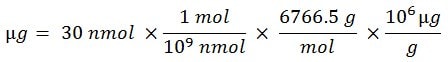
How do I conjugate dyes to amine-labeled sequences?
Please see our Protocol for Conjugating NHS-Ester Modifications to Amino-Labeled Oligonucleotides.
How do I reduce thiol-modified sequences?
Please see our Protocol for Thiol-Modified Oligonucleotide Reduction.
What are the MIQE guidelines for qPCR, and why should I follow them?
The MIQE guidelines are an attempt to standardize qPCR research to promote experimental transparency, ensure consistency between laboratories, and to maintain the integrity of the scientific literature. Details and a downloadable checklist can be found in our MIQE – Minimum Information for Publication of Quantitative Real-Time PCR Experiments article. You can view our qPCR and MIQE Seminar Series for additional details.
OLIGO STABILITY
If I left my oligonucleotides on the laboratory bench over the weekend, will they still work on Monday?
Yes, oligonucleotides, whether dry or in TE buffer, are typically stable for 3 - 6 months at room temperature. Details on stability can be found in our Oligonucleotide Handling & Stability article.
What are the recommended storage conditions?
We recommend keeping oligonucleotides at 4 °C for short-term use (stable for approximately 1 year) and at -20 °C for long-term storage (stable for approximately 2 years). Oligonucleotides can be kept dry or in solution (water or TE buffer) with no effect on stability. Details on stability can be found in our Oligonucleotide Handling & Stability article.
SAFETY DATA SHEET
Do you have safety data sheets available for your products?
Yes, please see our Safety Data Sheets page for downloadable PDFs.
GENERAL
Please visit KiCqStart® SYBR® Green Primers, KiCqStart® SYBR® Green Primers Gene Arrays, and KiCqStart® Probe Assays for specific FAQ on these products.
Do you have a frequently asked question that you think should be on this page? Please submit it to dnaoligos@sial.com.
To continue reading please sign in or create an account.
Don't Have An Account?