919381
CCW16
≥95%
Synonym(s):
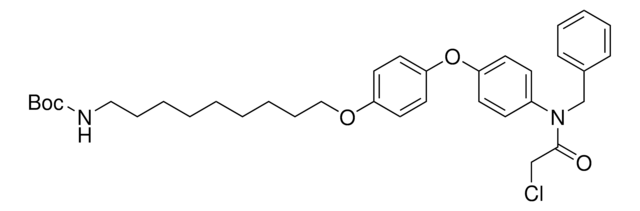
N-Benzyl-2-chloro-N-(4-(4-methoxyphenoxy)phenyl)acetamide, E3 ubiquitin ligase ligand, Ligand for PROTAC® research, RNF4-targeting ligand
About This Item
Recommended Products
ligand
CCW16
Quality Level
Assay
≥95%
form
powder
reaction suitability
reagent type: ligand
functional group
amine
storage temp.
2-8°C
SMILES string
COC1=CC=C(OC2=CC=C(N(CC3=CC=CC=C3)C(CCl)=O)C=C2)C=C1
Application
Other Notes
Portal: Building PROTAC® Degraders for Targeted Protein Degradation
Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications
Targeted Protein Degradation by Small Molecules
Small-Molecule PROTACS: New Approaches to Protein Degradation
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Legal Information
related product
Signal Word
Warning
Hazard Statements
Precautionary Statements
Hazard Classifications
Aquatic Acute 1 - Aquatic Chronic 1
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Articles
Protein Degrader Building Blocks are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Protein Degrader Building Blocks are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Protein Degrader Building Blocks are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Protein Degrader Building Blocks are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service