MBD0047
Enterococcus faecium FISH probe – ATTO 488
Probe for fluorescence in situ hybridization (FISH), 20µM in water
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
UNSPSC Code:
41105500
NACRES:
NA.54
Recommended Products
Quality Level
technique(s)
FISH: suitable
fluorescence
λex 504 nm; λem 521 nm
shipped in
dry ice
storage temp.
−20°C
General description
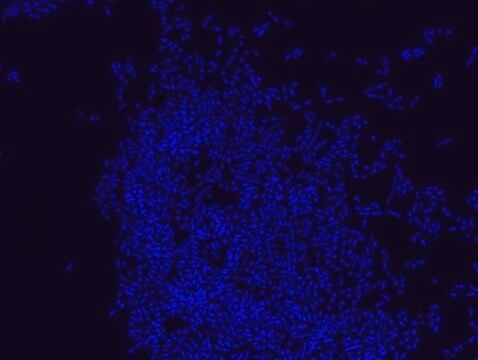
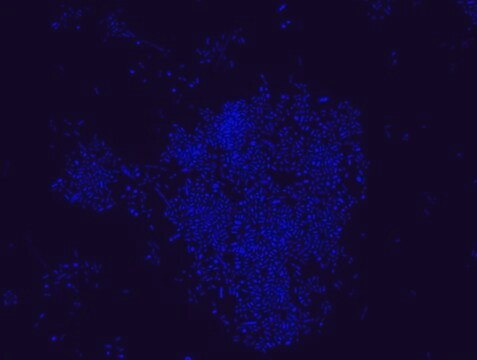
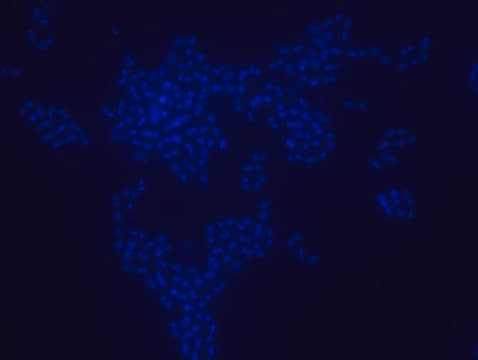
Fluorescent In Situ Hybridization technique (FISH) is based on the hybridization of fluorescent labeled oligonucleotide probe to a specific complementary DNA or RNA sequence in whole and intact cells. Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples. FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood and tissue), microbial ecology (solid biofilms, aquatic systems) and plants. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest. Enterococcus faecium probe specifically recognizes E. faecium cells,. E. faecium a gram-positive bacterium and is a cause of a variety of infections, including endocarditis, urinary tract infections, prostatitis, intra-abdominal infection, cellulitis, and wound infection as well as concurrent bacteremia. Studies have shown that E. faecium is highly resistant to multiple antibiotics and that vancomycin-resistant Enterococcus faecium (VRE) can be asymptomatically carried by healthy people, which can hamper hospitals’ attempts to control the spread of the bacteria. Over the past two decades, Enterococcus faecium has emerged as a leading cause of multidrug-resistant enterococcal infection in the United States. E. faecium has a high antibiotic-resistance with more than half of its pathogenic isolates expressing resistance to vancomycin, ampicillin, and high-levels of aminoglycosides.
Application
Probe for fluorescence in situ hybridization (FISH), recognizes Enterococcus faecium cells
Features and Benefits
- Visualize, identify, and isolate E. faecium cells.
- Observe native E. faecium cell populations in diverse microbiome environments.
- Specific, sensitive, and robust identification of E. faecium in bacterial mixed population.
- Specific, sensitive, and robust identification even when E. faecium is in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on E. faecium morphology and allows to study biofilm architecture.
- Identify E. faecium in clinical samples such as tumor and brain tissues (for example in formalin-fixed paraffin-embedded (FFPE) samples), saliva and oral cavity and medical equipment such as dental implants and voice prostheses.
- The ability to detect E. faecium in its natural habitat is an essential tool for studying host-microbiome interaction.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Rapid identification of clinically relevant Enterococcus species by fluorescence in situ hybridization
Wellinghausen, N., Bartel, M., Essig, A. & Poppert
Journal of Clinical Microbiology, 45, 3424?3426-3424?3426 (2007)
Fluorescent in situ hybridization with specific DNA probes offers adequate detection of Enterococcus faecalis and Enterococcus faecium in clinical samples
Waar, K., Degener, J. E., Van Luyn, M. J. & Harmsen, H. J. M.
Journal of Medical Microbiology, 54, 937?944-937?944 (2005)
NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007
Hidron, A. I. et al
Infection Control and Hospital Epidemiology : The Official Journal of the Society of Hospital Epidemiologists of America, 29, 996-1011 (2008)
Rapid identification of pathogens in blood cultures with a modified fluorescence in situ hybridization assay
Peters, R. P. H. et al.
Journal of Clinical Microbiology, 44, 4186-4188 (2066)
Genotyping and preemptive isolation to control an outbreak of vancomycin-resistant Enterococcus faecium
Mascini, E. M. et al.
Clinical Infectious Diseases, 42, 739-746 (2006)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service