Degrader Building Blocks for Targeted Protein Degradation
Targeted Protein Degradation
Targeted protein degradation is a novel strategy that uses small molecules to hijack endogenous proteolysis systems to degrade disease-relevant proteins. Reducing protein abundance in cells, phenotypes similar to gene-editing approaches (e.g. CRISPR-Cas9) can be achieved but with the advantages that come with small molecules. Not only is this of interest as a research tool to study the impact of selective protein knockdowns, but it has quickly been adopted by the global drug discovery community for its advantages over occupancy-based inhibition and drugging ~80% of proteins traditionally intractable to small molecules. Agents used in these approaches are called protein degraders, such as proteolysis-targeting chimeras (PROTAC® degraders) or molecular glues.
Heterobifunctional protein degraders contain a target-binding warhead on one end and an E3 ubiquitin ligase-targeting ligand on the other, connected by a linker in the middle (Figure 1). When the degrader is applied, it recruits the target to the E3 ligase. Once in proximity of the E3 ligase, the target is polyubiquitinated, flagging it for degradation via the proteasome.1-3
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.

Figure 1.Targeted protein degradation via proteolysis-targeting chimeras (PROTACs)
Challenges for Protein Degrader Synthesis
The design of degraders is challenging as slight alterations in structure can alter ternary complex formation and subsequent degradation.4 The 3D model in Figure 2 based on PDB 5T35 illustrates the importance of careful design to achieve binding to two disparate proteins (the target and E3 ligase) and the establishment of a protein–protein interface. Even with advances in computational chemistry, degrader design is still largely an empirical process where researchers generate libraries of degraders, taking a modular approach to varying the ligands, linkers, and exit vectors, an intense upfront chemistry endeavor.4,6

Figure 2.a) Chemical structure of representative PROTAC MZ1 highlighting target ligand, E3 ligase ligand, linker, and exit vector. b) Degrader structure impacts ternary complex formation, illustrated with MZ16. c) Empirical design requires the generation of libraries to test on case-by-case basis.
Streamlined Synthesis of Heterobifunctional Degrader Libraries
Our protein degrader building blocks are the easiest way to generate heterobifunctional degrader screening libraries from one starting target ligand to expedite degrader hit discovery. Within this building block collection that comprises all the components to construct degraders, our ligand–linker conjugates eliminate upfront synthetic steps, requiring only the chemistry to link a target ligand on the terminal functional group (Figure 3). Moreover, if the same terminal chemistry is selected, a chemist can simultaneously react 50+ ligand–linker conjugates with one starting target ligand in parallel to generate an initial screening library.

Figure 3.Ligand–linker conjugates
Diversity of the Ligand - Linker Conjugates
Our suite of ligand–linker conjugates contains strategic combinations of E3 ligands, exit vectors, linkers, and terminal chemistry.
E3 ligase recruiters and ligands: While more E3 ligases are being researched for targeted protein degradation, a handful are used most often in the development of protein degraders.7 Our conjugates include ligands and varied exit vectors for the validated E3 ligases CRBN, VHL, IAP, and RNF4 (Figure 4).
Linkers: Alkyl and PEG linkers are excellent starters to sample a range of hydrophobicity, flexibility, and lengths. In addition, we offer many “mixed”8 and rigid9–11 linkers to achieve diverse linker properties in your library (Figure 3).
Terminal chemistry: A variety of popular functional groups are available for linking the target warhead; our largest group includes terminal amines (Figure 3).
Advantages
- Synthetic time-saver: Ligand–linker conjugates simplify synthesis of single degraders and parallel synthesis for library construction
- Molecule design: Permutations of highest-interest E3 ligands, exit vectors, and linkers within the conjugates ease upfront combinatorial library design
- Compatibility: Linkers conjugate to common functional groups present on target ligands
- SAR: Strategic component variation built into the ligand–linker conjugates provides an upfront glimpse at SAR for informed optimization
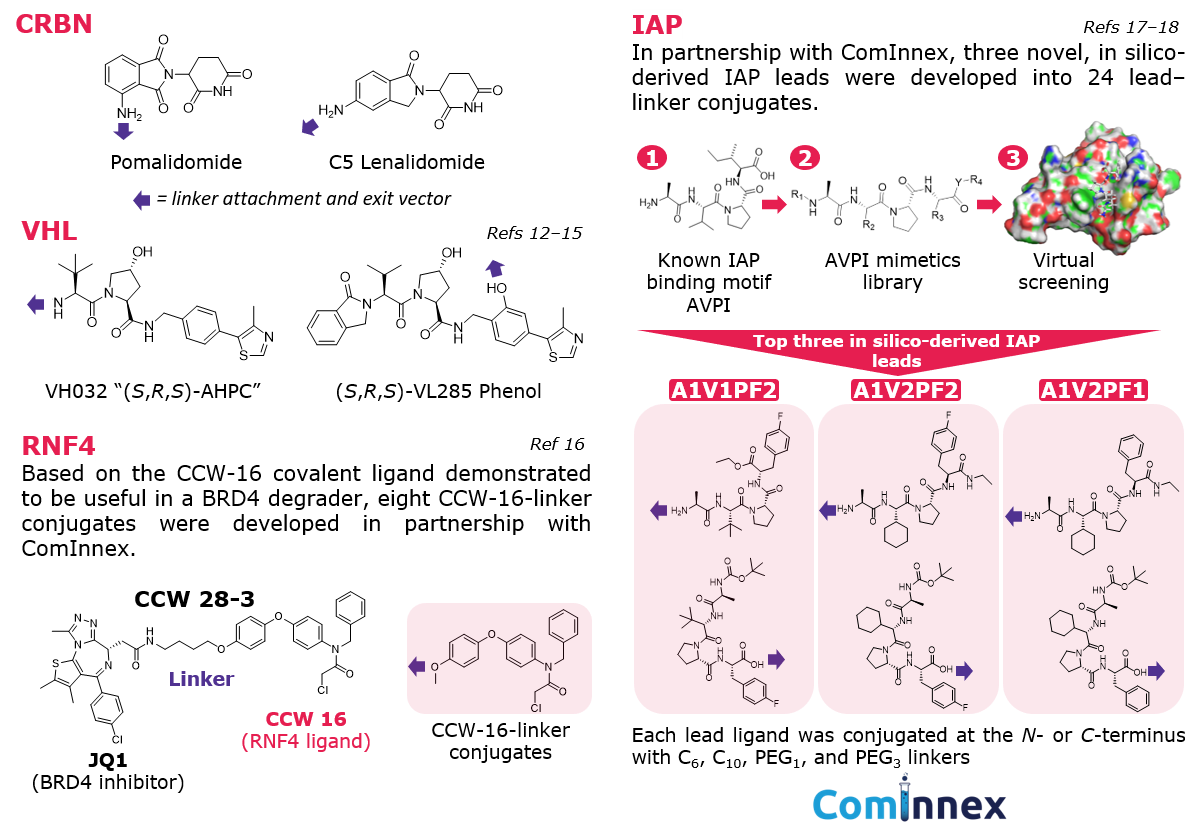
Figure 4.E3 Ligase ligands featured in conjugates
LET US HELP YOU BUILD YOUR DEGRADER LIBRARY BASED ON WHAT IS MOST IMPORTANT TO YOU.
Reach out to your MilliporeSigma technical specialist or SigmaAldrich.com/techservice for a sortable list or structure data file of all synthesis products, including 150+ ligand–linker conjugates, 250+ heterobifunctional linkers, 20+ ligands, and related probe compounds.
Related Articles
- Degrader Building Blocks with IAP In Silico-Derived Ligands
- Accelerating IAP-Based Protein Degrader Discovery with Novel Ligand Library
Webinars
References
Pour continuer à lire, veuillez vous connecter à votre compte ou en créer un.
Vous n'avez pas de compte ?