How does Protein AQUA work?
In June 2003, Dr. Steve Gygi and his team presented an innovative strategy, Protein AQUA™, enabling absolute protein quantitation using stable isotope labeled peptides and HPLC-MS. By applying a common principle, the use of a labeled molecule as an internal standard, to protein analysis, Gygi’s team has advanced the abilities of protein researchers to study complex biological samples quantitatively and has provided a valuable new tool for Proteomics.
An AQUA™ Peptide is simply a synthetic tryptic peptide corresponding to a peptide of interest. Each AQUA™ peptide incorporates one stable isotope labeled amino acid, creating a slight increase (6-10 daltons) in molecular weight. When mixed, the native peptide and the synthetic AQUA™ Peptide elute together chromatographically, migrate together electrophoreticly, and ionize with the same intensity. However, by mass spectrometry, the native peptide and the synthetic AQUA™ Peptide are easily distinguished.
In a typical AQUA™ experiment, a known amount of AQUA™ Peptide is added to a biological protein sample. The sample is then digested and analyzed by HPLC-MS. Extracted ion chromatograms are generated for the native peptide and the synthetic AQUA™ Peptide internal standard. Using peak ratios, the quantity of native peptide is calculated.
Protein-AQUA™ is a powerful, enabling technology, the limits of which are only now being explored. For proteomics researchers, it facilitates focused, quantitative studies of not only specific protein expression, but specific amino acid modification as well.
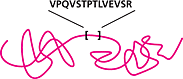
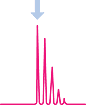
Optimize your LC-MS/MS protocol to resolve and monitor native and corresponding AQUA™ peptides

Add a known amount of AQUA™ Peptide to your biological protein sample
Digest with trypsin
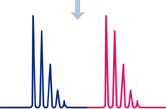
Analyze by LC-MS/MS. Generate extracted ion chromatograms for native and AQUA™ peptides, and calculate the amount of native peptide present.
To continue reading please sign in or create an account.
Don't Have An Account?